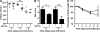Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM - PubMed (original) (raw)
Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM
Frédéric A Carvalho et al. J Exp Med. 2009.
Abstract
Abnormal expression of CEACAM6 is observed at the apical surface of the ileal epithelium in Crohn's disease (CD) patients, and CD ileal lesions are colonized by pathogenic adherent-invasive Escherichia coli (AIEC). We investigated the ability of AIEC reference strain LF82 to colonize the intestinal mucosa and to induce inflammation in CEABAC10 transgenic mice expressing human CEACAMs. AIEC LF82 virulent bacteria, but not nonpathogenic E. coli K-12, were able to persist in the gut of CEABAC10 transgenic mice and to induce severe colitis with reduced survival rate, marked weight loss, increased rectal bleeding, presence of erosive lesions, mucosal inflammation, and increased proinflammatory cytokine expression. The colitis depended on type 1 pili expression by AIEC bacteria and on intestinal CEACAM expression because no sign of colitis was observed in transgenic mice infected with type 1 pili-negative LF82-Delta fimH isogenic mutant or in wild-type mice infected with AIEC LF82 bacteria. These findings strongly support the hypothesis that in CD patients having an abnormal intestinal expression of CEACAM6, AIEC bacteria via type 1 pili expression can colonize the intestinal mucosa and induce gut inflammation. Thus, targeting AIEC adhesion to gut mucosa represents a new strategy for clinicians to prevent and/or to treat ileal CD.
Figures
Figure 1.
AIEC LF82 bacterial colonization and persistence in mouse intestine according to CEACAM expression. (A and B) Quantification of AIEC LF82 (A) or E. coli K-12 MG1655 (B) in the feces of WT (wt, white triangles) or CEABAC10 (tg, black triangles) mice receiving 0.25% DSS in drinking water after oral infection with 109 bacteria at day 0. (C) Quantification of colonic mucosal-associated AIEC LF82 bacteria at day 7 after oral infection. The quantification for each mouse (symbols) and the median (bars) is expressed as CFU/g of feces or CFU/mg of organ tissue. (D) Confocal microscopy analysis of colonic section at day 7 after infection from WT or transgenic mice infected with AIEC LF82. CEACAM6 expression was detected using anti-CEACAM6 monoclonal antibody and a Cy3-conjugated anti–mouse IgG. AIEC LF82 bacteria were detected using anti-O83 rabbit antibody and a FITC-conjugated anti–rabbit IgG. Arrow shows colocalization (yellow staining) between CEACAM6 and bacteria. WT FVB/N mice: E. coli K-12 MG1655 infected (n = 13), AIEC LF82 infected (n = 9); CEABAC10 transgenic FVB/N mice: E. coli K-12-MG1655 (n = 13), AIEC LF82-infected (n = 15). Results shown here are representative of three separate colonization experiments. *, P < 0.05; ***, P < 0.001. Bars, 50 µm.
Figure 2.
AIEC LF82 bacteria induced clinical symptoms of colitis in CEABAC10 transgenic mice. (A–F) AIEC LF82 infections were performed in WT (A–C) and in CEABAC10 (D–F) mice. Shown is evolution of body weight (A and D), survival rate (B and E), and DAI score ascertained at day 7 after infection (C and F) of mice orally challenged at day 0 with CMC alone (black circles), with CMC containing 109 E. coli K-12 MG1655 bacteria (white triangles), or with CMC containing 109 AIEC LF82 bacteria (white squares). (G) Quantification of AIEC LF82 at day 7 after infection in the liver and spleen. WT FVB/N mice: CMC alone (n = 11), E. coli K-12 MG1655 infected (n = 7), AIEC LF82 infected (n = 14); CEABAC10 transgenic FVB/N mice: CMC alone (n = 9), E. coli K-12 MG1655 infected (n = 11), AIEC LF82 infected (n = 15). Shown here are representative body weight evolution, survival rate, and DAI of three separate experiments. Horizontal bars represent medians. Error bars represent SEM. **, P < 0.01; ***, P < 0.001 (compared with noninfected mice).
Figure 3.
Intraperitoneal administration of anti-CEACAM6 monoclonal antibody 24 h before infection and at days 0, 1, and 3 after infection prevents colonization and clinical symptoms of colitis in AIEC LF82-infected CEABAC10 transgenic mice. (A–C) Evolution of the number of AIEC LF82 in the feces (A), of DAI score ascertained at days 3 and 6 after infection (B), and of body weight (C) of transgenic mice orally challenged at day 0 with 109 AIEC LF82 bacteria. Horizontal bars represent medians. CEABAC10 transgenic FVB/N mice: AIEC LF82 infected (n = 5; black triangles); AIEC LF82 infected after intraperitoneal injection of anti-CEACAM6 antibody (n = 5; black circles). Two experiments were performed independently. Shown here are colonization, DAI, and body weight evolution of one representative experiment. Error bars represent SEM. *, P < 0.05; **, P < 0.01.
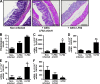
Figure 4.
Intestinal colonization by AIEC LF82 bacteria causes severe histopathological damage in colonic mucosa. (A–D) Hematoxylin/eosin/safran staining of colonic tissue sections obtained at day 7 after infection from WT mice (A) or CEABAC10 transgenic mice (C), noninfected or infected with E. coli K-12 MG1655 or with AIEC LF82 bacteria (magnification, 100×). Histopathological scoring for several parameters of colonic inflammation was performed for WT mice (B) or CEABAC10 transgenic mice (D), noninfected or infected with 109 E. coli K-12 MG1655 or with AIEC LF82 bacteria. WT FVB/N mice: CMC alone (n = 11), E. coli K-12 MG1655 infected (n = 12), AIEC LF82 infected (n = 14); CEABAC10 transgenic FVB/N mice: CMC alone (n = 9), E. coli K-12 MG1655 infected (n = 11), AIEC LF82 infected (n = 15). Shown here are representative histological scores of two separate experiments. Error bars represent SEM. ***, P < 0.001 compared with noninfected mice. Bars, 100 µm.
Figure 5.
AIEC LF82 colonization via type 1 pili expression induce clinical symptoms of colitis in CEABAC10 transgenic mice. (A) Quantification of AIEC LF82 bacteria (black triangles) or type 1 pili–negative AIEC LF82-Δ_fimH_ mutant (white triangles) in the feces of CEABAC10 mice after oral infection with 109 bacteria at day 0. Each symbol represents one mouse and the horizontal bars represent the medians. (B and C) Evolution of body weight (B) and survival rate (C) of CEABAC10 transgenic mice orally challenged at day 0 with CMC alone (black circles), CMC containing 109 AIEC LF82 bacteria (white squares), or CMC containing 109 AIEC LF82-Δ_fimH_ bacteria (white triangles). (D) DAI score ascertained for CEABAC10 transgenic mice that were noninfected, infected with AIEC LF82-Δ_fimH_ bacteria, or infected with AIEC LF82 bacteria at day 7 after infection. CEABAC10 transgenic FVB/N mice: CMC alone (n = 9), AIEC LF82-Δ_fimH_ infected (n = 14), AIEC LF82 infected (n = 15). Shown here are representative body weight evolution, survival rate, and DAI of three separate experiments. Error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with mice infected with AIEC LF82-Δ_fimH_).
Figure 6.
Type 1 pili–dependent AIEC LF82 colonization is required to induce severe histopathological damage and inflammation of colonic mucosa. (A) Hematoxylin/eosin/safran staining of colonic tissue sections obtained at day 7 after infection from CEABAC10 transgenic mice noninfected, infected with AIEC LF82-Δ_fimH_ isogenic mutant, or infected with AIEC LF82 bacteria (magnification, 100×). (B) Histopathological scoring for several parameters of colonic inflammation. (C–F) Total RNAs from mouse colon were isolated and IL-1β (C), IL-6 (D), IL-17 (E), and IL-10 (F) mRNA levels were measured by RT-PCR. CEABAC10 transgenic FVB/N mice: CMC alone (n = 9), AIEC LF82-Δ_fimH_ infected (n = 14), AIEC LF82 infected (n = 15). Shown here are representative histological scores of two separate experiments. Error bars represent SEM. *, P < 0.05; **, P < 0.01 (compared with mice infected with AIEC LF82-Δ_fimH_). Bars, 100 µm.
Similar articles
- Development of Heptylmannoside-Based Glycoconjugate Antiadhesive Compounds against Adherent-Invasive Escherichia coli Bacteria Associated with Crohn's Disease.
Sivignon A, Yan X, Alvarez Dorta D, Bonnet R, Bouckaert J, Fleury E, Bernard J, Gouin SG, Darfeuille-Michaud A, Barnich N. Sivignon A, et al. mBio. 2015 Nov 17;6(6):e01298-15. doi: 10.1128/mBio.01298-15. mBio. 2015. PMID: 26578673 Free PMC article. - Point mutations in FimH adhesin of Crohn's disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response.
Dreux N, Denizot J, Martinez-Medina M, Mellmann A, Billig M, Kisiela D, Chattopadhyay S, Sokurenko E, Neut C, Gower-Rousseau C, Colombel JF, Bonnet R, Darfeuille-Michaud A, Barnich N. Dreux N, et al. PLoS Pathog. 2013 Jan;9(1):e1003141. doi: 10.1371/journal.ppat.1003141. Epub 2013 Jan 24. PLoS Pathog. 2013. PMID: 23358328 Free PMC article. - Saccharomyces cerevisiae CNCM I-3856 prevents colitis induced by AIEC bacteria in the transgenic mouse model mimicking Crohn's disease.
Sivignon A, de Vallée A, Barnich N, Denizot J, Darcha C, Pignède G, Vandekerckove P, Darfeuille-Michaud A. Sivignon A, et al. Inflamm Bowel Dis. 2015 Feb;21(2):276-86. doi: 10.1097/MIB.0000000000000280. Inflamm Bowel Dis. 2015. PMID: 25569734 - E. coli-mediated gut inflammation in genetically predisposed Crohn's disease patients.
Barnich N, Denizot J, Darfeuille-Michaud A. Barnich N, et al. Pathol Biol (Paris). 2013 Oct;61(5):e65-9. doi: 10.1016/j.patbio.2010.01.004. Epub 2010 Apr 8. Pathol Biol (Paris). 2013. PMID: 20381273 Review. - Adherent-invasive Escherichia coli in inflammatory bowel disease.
Palmela C, Chevarin C, Xu Z, Torres J, Sevrin G, Hirten R, Barnich N, Ng SC, Colombel JF. Palmela C, et al. Gut. 2018 Mar;67(3):574-587. doi: 10.1136/gutjnl-2017-314903. Epub 2017 Nov 15. Gut. 2018. PMID: 29141957 Review.
Cited by
- An adherent-invasive Escherichia coli-colonized mouse model to evaluate microbiota-targeting strategies in Crohn's disease.
Sivignon A, Chervy M, Chevarin C, Ragot E, Billard E, Denizot J, Barnich N. Sivignon A, et al. Dis Model Mech. 2022 Oct 1;15(10):dmm049707. doi: 10.1242/dmm.049707. Epub 2022 Oct 24. Dis Model Mech. 2022. PMID: 36172858 Free PMC article. - Preventive Effect of Spontaneous Physical Activity on the Gut-Adipose Tissue in a Mouse Model That Mimics Crohn's Disease Susceptibility.
Maillard F, Vazeille E, Sauvanet P, Sirvent P, Bonnet R, Combaret L, Chausse P, Chevarin C, Otero YF, Delcros G, Chavanelle V, Boisseau N, Barnich N. Maillard F, et al. Cells. 2019 Jan 9;8(1):33. doi: 10.3390/cells8010033. Cells. 2019. PMID: 30634469 Free PMC article. - Intestinal homeostasis and inflammation: Gut microbiota at the crossroads of pancreas-intestinal barrier axis.
Zhang Z, Tanaka I, Pan Z, Ernst PB, Kiyono H, Kurashima Y. Zhang Z, et al. Eur J Immunol. 2022 Jul;52(7):1035-1046. doi: 10.1002/eji.202149532. Epub 2022 May 15. Eur J Immunol. 2022. PMID: 35476255 Free PMC article. Review. - Dietary L-serine confers a competitive fitness advantage to Enterobacteriaceae in the inflamed gut.
Kitamoto S, Alteri CJ, Rodrigues M, Nagao-Kitamoto H, Sugihara K, Himpsl SD, Bazzi M, Miyoshi M, Nishioka T, Hayashi A, Morhardt TL, Kuffa P, Grasberger H, El-Zaatari M, Bishu S, Ishii C, Hirayama A, Eaton KA, Dogan B, Simpson KW, Inohara N, Mobley HLT, Kao JY, Fukuda S, Barnich N, Kamada N. Kitamoto S, et al. Nat Microbiol. 2020 Jan;5(1):116-125. doi: 10.1038/s41564-019-0591-6. Epub 2019 Nov 4. Nat Microbiol. 2020. PMID: 31686025 Free PMC article. - Activation of the EIF2AK4-EIF2A/eIF2α-ATF4 pathway triggers autophagy response to Crohn disease-associated adherent-invasive Escherichia coli infection.
Bretin A, Carrière J, Dalmasso G, Bergougnoux A, B'chir W, Maurin AC, Müller S, Seibold F, Barnich N, Bruhat A, Darfeuille-Michaud A, Nguyen HT. Bretin A, et al. Autophagy. 2016 May 3;12(5):770-83. doi: 10.1080/15548627.2016.1156823. Epub 2016 Mar 17. Autophagy. 2016. PMID: 26986695 Free PMC article.
References
- Barnich N., Boudeau J., Claret L., Darfeuille-Michaud A. 2003. Regulatory and functional co-operation of flagella and type 1 pili in adhesive and invasive abilities of AIEC strain LF82 isolated from a patient with Crohn's disease.Mol. Microbiol. 48:781–794 doi:<10.1046/j.1365-2958.2003.03468.x> - DOI - PubMed
- Barnich N., Carvalho F.A., Glasser A.L., Darcha C., Jantscheff P., Allez M., Peeters H., Bommelaer G., Desreumaux P., Colombel J.F., Darfeuille-Michaud A. 2007. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease.J. Clin. Invest. 117:1566–1574 doi:<10.1172/JCI30504> - DOI - PMC - PubMed
- Baumgart M., Dogan B., Rishniw M., Weitzman G., Bosworth B., Yantiss R., Orsi R.H., Wiedmann M., McDonough P., Kim S.G., et al. 2007. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum.ISME J. 1:403–418 doi:<10.1038/ismej.2007.52> - DOI - PubMed
- Boudeau J., Glasser A.L., Julien S., Colombel J.F., Darfeuille-Michaud A. 2003. Inhibitory effect of probiotic Escherichia coli strain Nissle 1917 on adhesion to and invasion of intestinal epithelial cells by adherent-invasive E. coli strains isolated from patients with Crohn's disease.Aliment. Pharmacol. Ther. 18:45–56 doi:<10.1046/j.1365-2036.2003.01638.x> - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases