Structural basis for phototoxicity of the genetically encoded photosensitizer KillerRed - PubMed (original) (raw)
Structural basis for phototoxicity of the genetically encoded photosensitizer KillerRed
Sergei Pletnev et al. J Biol Chem. 2009.
Abstract
KillerRed is the only known fluorescent protein that demonstrates notable phototoxicity, exceeding that of the other green and red fluorescent proteins by at least 1,000-fold. KillerRed could serve as an instrument to inactivate target proteins or to kill cell populations in photodynamic therapy. However, the nature of KillerRed phototoxicity has remained unclear, impeding the development of more phototoxic variants. Here we present the results of a high resolution crystallographic study of KillerRed in the active fluorescent and in the photobleached non-fluorescent states. A unique and striking feature of the structure is a water-filled channel reaching the chromophore area from the end cap of the beta-barrel that is probably one of the key structural features responsible for phototoxicity. A study of the structure-function relationship of KillerRed, supported by structure-based, site-directed mutagenesis, has also revealed the key residues most likely responsible for the phototoxic effect. In particular, Glu(68) and Ser(119), located adjacent to the chromophore, have been assigned as the primary trigger of the reaction chain.
Figures
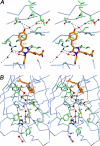
FIGURE 1.
The environment of the chromophore in KillerRed. A, stereoview of a chain of hydrogen-bonded waters (red spheres in the upper part of the drawing) flowing from the outside to the phenolic ring hydroxyl of the QYG chromophore (shown in orange) through a pore located on the cylindrical β-barrel surface composed of the backbone of residues 142–144 and 199–201. A fraction of the other water chain (lower part of the drawing; see the legend for B) and the key residues Asn145 and Thr201 and the catalytic Arg94 and Glu218, interacting with chromophore, are also shown. B, stereoview of the water-filled channel with a chain of seven consecutive hydrogen-bonded water molecules (red spheres) traversing along the β-barrel from Pro192 at the cap loop to the catalytic Glu218 in the chromophore area. Two other water molecules are shown at the assumed entry point outside of the β-barrel (figure created with SETOR (55)).
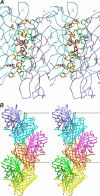
FIGURE 2.
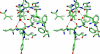
Oligomerization of KillerRed. A, stereoview of the interface between the A and B subunits in the KillerRed dimer that are related by a 2-fold non-crystallographic symmetry axis. B, KillerRed oligomeric assembly in the crystal lattice showing three consecutive dimers composed from the β-barrel monomeric units (figure created with SETOR (55)).
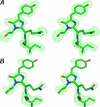
FIGURE 3.
Stereoviews of the QYG chromophore with a preceding Ile residue in the weighted omit mFo − DFc electron density map (18) (cut-off ρ = 3.0σ). A, active fluorescent state; B, photobleached non-fluorescent state after 5 h of green laser light irradiation at λ = 532 nm. The latter density indicates considerable damage to the Gln65 and Tyr66 side chains of the chromophore (figure created with PyMOL (56)).
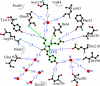
FIGURE 4.
A schematic diagram illustrating the environment of the chromophore in the KillerRed structure. Hydrogen bonds (≤3.3 Å) are shown as blue dashed lines, water molecules (W) are shown as red spheres, and van der Waals contacts (≤3.9 Å) are shown as black “_eyelashes_” (figure prepared with LIGPLOT/HBPLUS (57)).
FIGURE 5.
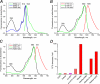
Spectral transitions of KillerRed in solution. A, reduction of red fluorescence intensity of KillerRed immobilized on beads upon the addition of 100 m
m
βME and its recovery after βME has been washed out. B, normalized photobleaching curves for KillerRed immobilized on beads in the absence and presence of βME upon excitation by mercury arc lamp (540–580 nm, 0.2 watt/cm2). C, absorbance spectrum changes in the presence of βME at different concentrations (shown in m
m
above each curve). An inset shows the dependence of absorbance at 585 nm on βME concentration. D, absorbance spectrum changes during photobleaching with a green laser at 532 nm.
FIGURE 6.
The key residues adjacent to the chromophore. Glu68 is hydrogen-bonded via two waters to Ser119, in a mostly hydrophobic pocket. The side chain of Ser119 shows two alternative orientations (created with SETOR (55)).
FIGURE 7.
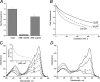
Absorption, excitation, and emission spectra of the mutants of KillerRed. A, KillerRed-E68Q; B, KillerRed-S119A; C, KillerRed-A220T. D, decrease of viable bacterial counts (in colony-forming units (CFU)) after white light illumination of E. coli expressing the designated fluorescent proteins.
FIGURE 8.
Structure-based sequence alignment of KillerRed, its wild-type progenitor anm2CP, DsRed, and avGFP. The numbering of residues in KillerRed and avGFP is given above and below the corresponding sequences. Buried residues are highlighted in gray. The chromophore-forming sequence is underlined. In red are shown residues residing in the nearest environment of the chromophore (≤3.9 Å); among them are the catalytic residues, Arg94 and Glu218 (highlighted in yellow), whereas the KillerRed/anm2CP differences are highlighted in blue.
FIGURE 9.
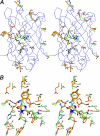
A comparison of KillerRed with other FPs. A, stereoview showing the distribution of the amino acid differences (see Fig. 8) between the phototoxic, fluorescent KillerRed (element C in green) and the non-toxic, non-fluorescent anm2CP (element C in orange). B, the neighborhood of the chromophore in the superimposed three-dimensional structures of KillerRed (element C in green) and DsRed (element C in orange) (created with SETOR (55)).
Similar articles
- Crystal Structure of Phototoxic Orange Fluorescent Proteins with a Tryptophan-Based Chromophore.
Pletneva NV, Pletnev VZ, Sarkisyan KS, Gorbachev DA, Egorov ES, Mishin AS, Lukyanov KA, Dauter Z, Pletnev S. Pletneva NV, et al. PLoS One. 2015 Dec 23;10(12):e0145740. doi: 10.1371/journal.pone.0145740. eCollection 2015. PLoS One. 2015. PMID: 26699366 Free PMC article. - Phototoxic effects of lysosome-associated genetically encoded photosensitizer KillerRed.
Serebrovskaya EO, Ryumina AP, Boulina ME, Shirmanova MV, Zagaynova EV, Bogdanova EA, Lukyanov SA, Lukyanov KA. Serebrovskaya EO, et al. J Biomed Opt. 2014 Jul;19(7):071403. doi: 10.1117/1.JBO.19.7.071403. J Biomed Opt. 2014. PMID: 24365992 - A proton transfer network that generates deprotonated tyrosine is a key to producing reactive oxygen species in phototoxic KillerRed protein.
Lee W , Kim I , Rhee YM . Lee W , et al. Phys Chem Chem Phys. 2018 Aug 29;20(34):22342-22350. doi: 10.1039/c8cp02939c. Phys Chem Chem Phys. 2018. PMID: 30128469 - Advances in the Genetically Engineered KillerRed for Photodynamic Therapy Applications.
Liu J, Wang F, Qin Y, Feng X. Liu J, et al. Int J Mol Sci. 2021 Sep 20;22(18):10130. doi: 10.3390/ijms221810130. Int J Mol Sci. 2021. PMID: 34576293 Free PMC article. Review. - [Chromoproteins of the green fluorescent protein family: properties and applications].
Shkrob M, Mishin AS, Chudakov DM, Labas IuA, Luk'ianov KA. Shkrob M, et al. Bioorg Khim. 2008 Sep-Oct;34(5):581-90. doi: 10.1134/s1068162008050014. Bioorg Khim. 2008. PMID: 19060933 Review. Russian.
Cited by
- Chromophore-assisted laser inactivation in neural development.
Li W, Stuurman N, Ou G. Li W, et al. Neurosci Bull. 2012 Aug;28(4):333-41. doi: 10.1007/s12264-012-1252-4. Neurosci Bull. 2012. PMID: 22833033 Free PMC article. Review. - The application of KillerRed for acute protein inactivation in living cells.
Jarvela TS, Linstedt AD. Jarvela TS, et al. Curr Protoc Cytom. 2014 Jul 1;69:12.35.1-12.35.10. doi: 10.1002/0471142956.cy1235s69. Curr Protoc Cytom. 2014. PMID: 24984963 Free PMC article. - Optical inactivation of intracellular molecules by fast-maturating photosensitizing fluorescence protein, HyperNova.
Shidara H, Shirai T, Ozaki-Noma R, Jitsuki S, Nagai T, Takemoto K. Shidara H, et al. Commun Biol. 2024 Aug 6;7(1):945. doi: 10.1038/s42003-024-06583-x. Commun Biol. 2024. PMID: 39107369 Free PMC article. - Positive-Type Reversibly Photoswitching Red Fluorescent Protein for Dual-Color Superresolution Imaging with Single Light Exposure for Off-Switching.
Ozaki-Noma R, Wazawa T, Kakizuka T, Shidara H, Takemoto K, Nagai T. Ozaki-Noma R, et al. ACS Nano. 2025 Feb 25;19(7):7188-7201. doi: 10.1021/acsnano.4c16847. Epub 2025 Feb 12. ACS Nano. 2025. PMID: 39937184 Free PMC article. - Quantification of reactive oxygen species production by the red fluorescent proteins KillerRed, SuperNova and mCherry.
Onukwufor JO, Trewin AJ, Baran TM, Almast A, Foster TH, Wojtovich AP. Onukwufor JO, et al. Free Radic Biol Med. 2020 Feb 1;147:1-7. doi: 10.1016/j.freeradbiomed.2019.12.008. Epub 2019 Dec 10. Free Radic Biol Med. 2020. PMID: 31841676 Free PMC article.
References
- Verkhusha V. V., Lukyanov K. A. (2004) Nat. Biotechnol. 22, 289–296 - PubMed
- Chudakov D. M., Lukyanov S., Lukyanov K. A. (2005) Trends Biotechnol. 23, 605–613 - PubMed
- Zubova N. N., Savitsky A. P. (2005) Usp. Biol. Khim. 45, 1–66
- Seitz G., Warmann S. W., Fuchs J., Mau-Holzmann U. A., Ruck P., Heitmann H., Hoffman R. M., Mahrt J., Müller G. A., Wessels J. T. (2006) J. Pediatr. Surg. 41, 1369–1376 - PubMed
- Remington S. J. (2006) Curr. Opin. Struct. Biol. 16, 714–721 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHSN2612008000001E/PHS HHS/United States
- ImNIH/Intramural NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- R24 MD002780/MD/NIMHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous