Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex - PubMed (original) (raw)
Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex
Qun Zhao et al. J Biol Chem. 2009.
Abstract
Accessibility within chromatin is an important factor in the prompt removal of UV-induced DNA damage by nucleotide excision repair (NER). Chromatin remodeling by the SWI/SNF complex has been shown to play an important modulating role in NER in vitro and yeast in vivo. Nevertheless, the molecular basis of cross-talk between SWI/SNF and NER in mammalian cells is not fully understood. Here, we show that knockdown of Brg1, the ATPase subunit of SWI/SNF, negatively affects the elimination of cyclobutane pyrimidine dimers (CPD), but not of pyrimidine (6, 4)pyrimidone photoproducts (6-4PP) following UV irradiation of mammalian cells. Brg1-deficient cells exhibit a lower chromatin relaxation as well as impaired recruitment of downstream NER factors, XPG and PCNA, to UV lesions. However, the assembly of upstream NER factors, DDB2 and XPC, at the damage site was unaffected by Brg1 knockdown. Interestingly, Brg1 interacts with XPC within chromatin and is recruited to UV-damaged sites in a DDB2- and XPC-dependent manner. Also, postirradiation decrease of XPC levels occurred more rapidly in Brg1-deficient than normal cells. Conversely, XPC transcription remained unaltered upon Brg1 knockdown indicating that Brg1 affects the stability of XPC protein following irradiation. Thus, Brg1 facilitates different stages of NER by initially modulating UV-induced chromatin relaxation and stabilizing XPC at the damage sites, and subsequently stimulating the recruitment of XPG and PCNA to successfully culminate the repair.
Figures
FIGURE 1.
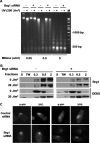
Brg1 is required for efficient removal of CPD but not 6-4PP. A, knockdown of Brg1 level by hBrg1 siRNA. OSU-2 cells were transfected with Brg1 siRNA (20 n
m
) using Lipofectamine 2000. Cell lysate was collected 24 or 48 h after transfection and subjected to Western blot analysis with anti-Brg1 antibody. Lamin B was detected to serve as an internal control. B and C, effect of Brg1 on the efficiency of CPD (B) or 6-4PP (C) removal. After 24 h of transfection with Brg1 siRNA, OSU-2 cells were cultured in serum-free medium for an additional 24 h. Cells were UV irradiated at 10 J/m2 and allowed to repair for indicated times. The same amount of genomic DNA was subjected to ISB analysis and the amount of CPD (B) or 6-4PP (C) was detected with anti-CPD or anti-6-4PP antibody, respectively, at each time point. Data presented in the graph on the right represent the relative CPD or 6-4PP remaining in each sample. Percentage CPD or 6-4PP was calculated from the intensity relative to initial irradiated sample. The data points represent an average of three independent measurements, with error bars representing standard deviation.
FIGURE 2.
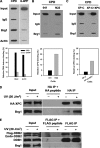
Brg1 is indispensable for UV-induced chromatin relaxation but has no effect on DDB2, XPC, and XPB recruitment to damage sites. A, knockdown of Brg1 affects UV-induced chromatin relaxation. OSU-2 cells were transfected with control siRNA or Brg1 siRNA for 24 h and serum starved for an additional 24 h and then UV irradiated at 200 J/m2. Cells were cultured in the same medium for 30 min, and nuclei were isolated and subjected to MNase digestion assay to determine the extent of chromatin relaxation. B, binding of Brg1 or DDB2 to UV-damaged chromatin. OSU-2 cells were transfected with control siRNA or Brg1 siRNA for 24 h and UV irradiated at 20 J/m2. Cells were incubated in the same medium for another 30 min and subjected to cellular protein fractionation. Five fractions were obtained according to the resistance to detergent and salt, e.g. S, cytoplasmic soluble protein; TW, nucleoplasmic soluble proteins; 0.3, proteins binding to chromatin loosely; 0.5, proteins binding to chromatin with intermediate affinity; 2.0, proteins binding to chromatin tightly. Each protein fraction, corresponding to an equivalent cell number, was analyzed by Western blotting with anti-Brg1 or anti-DDB2 antibody. C, recruitment of XPC or XPB to UV-damage. OSU-2 cells with or without Brg1 siRNA transfection were grown on coverslips and UV irradiated (40 J/m2) through a 5-μm isopore polycarbonate filter. Cells were incubated in medium for 30 min and then fixed with 2% paraformaldehyde. Cells were double stained with mouse anti-CPD antibody and rabbit anti-XPC antibody or rabbit anti-XPB antibody.
FIGURE 3.
Brg1 interacts with XPC, and its recruitment to CPD depends on both DDB2 and XPC. A, recruitment of Brg1 to UV-damage sites. OSU-2 cells were UV irradiated at 20 J/m2 and repaired for 30 min. Cells were fixed with 1% formaldehyde and chromatin was isolated and subjected to immunoprecipitation with anti-Brg1 or anti-AcH3 (Lys-9, 14) antibodies or normal rabbit IgG. An input sample represents 2% of the total chromatin subjected to ChIP. ISB was carried out with anti-CPD or anti-6-4PP antibody to determine the level of damage (CPD or 6-4PP) in the isolated chromatin. B, Brg1 recruitment to CPD depends on DDB2. 041 or N22 cells were treated with UV and processed as in A. C, Brg1 recruitment to CPD depends on XPC. XP-C or XP-C cells transiently transfected with XPC plasmid were treated and processed as in A. The IgG-nonspecific background was deducted from each specific band, and the relative amount of CPD or 6-4PP bound to Brg1 or AcH3 was then calculated relative to the respective input levels set as 100%. D, Brg1 interacts with XPC on the chromatin. HeLa-XPC cells were treated with UV at 20 J/m2 and allowed to repair for 30 min. Cell nuclei were isolated and digested with Benzonase nuclease to release chromatin-associated proteins and immunoprecipitated with anti-HA affinity gel. IP mixture was subjected to Western blot analysis with anti-Brg1 antibody. E, Brg1 does not interact with DDB2 on the chromatin. HeLa-DDB2 cells were treated with UV at 20 J/m2 and allowed to repair for 30 min. Cell nuclei were isolated and digested with Benzonase nuclease to release chromatin-associated proteins and immunoprecipitated with anti-FLAG M2 affinity gel. IP mixture was subjected to Western blot analysis with anti-Brg1 antibody.
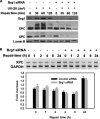
FIGURE 4.
Brg1 protects XPC from degradation after UV. A, Brg1 affects XPC protein levels in response to UV. OSU-2 cells were transfected with control siRNA or Brg1 siRNA for 48 h and treated with UV at 20 J/m2 and allowed to repair for indicated time periods. Cell lysates were subjected to Western blotting with anti-XPC antibody. Brg1 level was detected to check the efficiency of siRNA knockdown and Lamin B was used as an internal control. B, Brg1 did not affect transcription level of XPC before or after UV. OSU-2 cells were transfected with either control siRNA or Brg1 siRNA for 48 h and treated with UV at 20 J/m2 and further cultured for the indicated time periods. RT-PCR was used to analyze mRNA levels of XPC. GAPDH mRNA was used as an internal control for normalization between samples. Values are expressed as fold increase relative to the XPC mRNA levels without UV irradiation. Each point represents the mean of three determinations with error bars representing standard deviations.
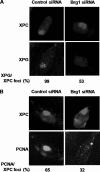
FIGURE 5.
Brg1 affects the recruitment of XPG and PCNA to UV damage sites. A, recruitment of XPG to DNA damage sites. OSU-2 cells were UV treated at 40 J/m2 through a filter containing pores of 5 μm in diameter and double stained with rabbit anti-XPC antibody and mouse anti-XPG antibody. B, OSU-2 cells were treated and processed as in A and double stained with rabbit anti-XPC antibody and mouse anti-PCNA antibody. The percentage of foci-containing cells were obtained from scoring >300 cells within a uniformly defined field.
Similar articles
- Downregulation of SWI/SNF chromatin remodeling factor subunits modulates cisplatin cytotoxicity.
Kothandapani A, Gopalakrishnan K, Kahali B, Reisman D, Patrick SM. Kothandapani A, et al. Exp Cell Res. 2012 Oct 1;318(16):1973-86. doi: 10.1016/j.yexcr.2012.06.011. Epub 2012 Jun 18. Exp Cell Res. 2012. PMID: 22721696 Free PMC article. - Human SNF5/INI1, a component of the human SWI/SNF chromatin remodeling complex, promotes nucleotide excision repair by influencing ATM recruitment and downstream H2AX phosphorylation.
Ray A, Mir SN, Wani G, Zhao Q, Battu A, Zhu Q, Wang QE, Wani AA. Ray A, et al. Mol Cell Biol. 2009 Dec;29(23):6206-19. doi: 10.1128/MCB.00503-09. Epub 2009 Oct 5. Mol Cell Biol. 2009. PMID: 19805520 Free PMC article. - The chromatin remodeling factor BRG1 stimulates nucleotide excision repair by facilitating recruitment of XPC to sites of DNA damage.
Zhang L, Zhang Q, Jones K, Patel M, Gong F. Zhang L, et al. Cell Cycle. 2009 Dec;8(23):3953-9. doi: 10.4161/cc.8.23.10115. Cell Cycle. 2009. PMID: 19901545 No abstract available. - Open, repair and close again: chromatin dynamics and the response to UV-induced DNA damage.
Palomera-Sanchez Z, Zurita M. Palomera-Sanchez Z, et al. DNA Repair (Amst). 2011 Feb 7;10(2):119-25. doi: 10.1016/j.dnarep.2010.10.010. Epub 2010 Dec 3. DNA Repair (Amst). 2011. PMID: 21130713 Review. - The SWI/SNF complex and cancer.
Reisman D, Glaros S, Thompson EA. Reisman D, et al. Oncogene. 2009 Apr 9;28(14):1653-68. doi: 10.1038/onc.2009.4. Epub 2009 Feb 23. Oncogene. 2009. PMID: 19234488 Review.
Cited by
- Molecular regulation of UV-induced DNA repair.
Shah P, He YY. Shah P, et al. Photochem Photobiol. 2015 Mar-Apr;91(2):254-64. doi: 10.1111/php.12406. Epub 2015 Jan 14. Photochem Photobiol. 2015. PMID: 25534312 Free PMC article. Review. - Multiple interaction nodes define the postreplication repair response to UV-induced DNA damage that is defective in melanomas and correlated with UV signature mutation load.
Pavey S, Pinder A, Fernando W, D'Arcy N, Matigian N, Skalamera D, Lê Cao KA, Loo-Oey D, Hill MM, Stark M, Kimlin M, Burgess A, Cloonan N, Sturm RA, Gabrielli B. Pavey S, et al. Mol Oncol. 2020 Jan;14(1):22-41. doi: 10.1002/1878-0261.12601. Epub 2019 Dec 19. Mol Oncol. 2020. PMID: 31733171 Free PMC article. - A chromatin scaffold for DNA damage recognition: how histone methyltransferases prime nucleosomes for repair of ultraviolet light-induced lesions.
Gsell C, Richly H, Coin F, Naegeli H. Gsell C, et al. Nucleic Acids Res. 2020 Feb 28;48(4):1652-1668. doi: 10.1093/nar/gkz1229. Nucleic Acids Res. 2020. PMID: 31930303 Free PMC article. - Regulation of the Mammalian SWI/SNF Family of Chromatin Remodeling Enzymes by Phosphorylation during Myogenesis.
Padilla-Benavides T, Reyes-Gutierrez P, Imbalzano AN. Padilla-Benavides T, et al. Biology (Basel). 2020 Jul 3;9(7):152. doi: 10.3390/biology9070152. Biology (Basel). 2020. PMID: 32635263 Free PMC article. Review. - ATP-dependent chromatin remodeling in the DNA-damage response.
Lans H, Marteijn JA, Vermeulen W. Lans H, et al. Epigenetics Chromatin. 2012 Jan 30;5:4. doi: 10.1186/1756-8935-5-4. Epigenetics Chromatin. 2012. PMID: 22289628 Free PMC article.
References
- de Laat W. L., Jaspers N. G., Hoeijmakers J. H. (1999) Genes Dev. 13, 768–785 - PubMed
- Hanawalt P. C. (2002) Oncogene 21, 8949–8956 - PubMed
- Sancar A. (1996) Annu. Rev. Biochem. 65, 43–81 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ES2388/ES/NIEHS NIH HHS/United States
- R01 ES012991/ES/NIEHS NIH HHS/United States
- R01 CA093413/CA/NCI NIH HHS/United States
- R01 ES002388/ES/NIEHS NIH HHS/United States
- ES12991/ES/NIEHS NIH HHS/United States
- CA93413/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous