De novo genome sequence assembly of a filamentous fungus using Sanger, 454 and Illumina sequence data - PubMed (original) (raw)
doi: 10.1186/gb-2009-10-9-r94. Epub 2009 Sep 11.
Nancy Y Liao, Darren Platt, Gordon Robertson, Michael Seidel, Simon K Chan, T Roderick Docking, Inanc Birol, Robert A Holt, Martin Hirst, Elaine Mardis, Marco A Marra, Richard C Hamelin, Jörg Bohlmann, Colette Breuil, Steven Jm Jones
Affiliations
- PMID: 19747388
- PMCID: PMC2768983
- DOI: 10.1186/gb-2009-10-9-r94
De novo genome sequence assembly of a filamentous fungus using Sanger, 454 and Illumina sequence data
Scott Diguistini et al. Genome Biol. 2009.
Abstract
Sequencing-by-synthesis technologies can reduce the cost of generating de novo genome assemblies. We report a method for assembling draft genome sequences of eukaryotic organisms that integrates sequence information from different sources, and demonstrate its effectiveness by assembling an approximately 32.5 Mb draft genome sequence for the forest pathogen Grosmannia clavigera, an ascomycete fungus. We also developed a method for assessing draft assemblies using Illumina paired end read data and demonstrate how we are using it to guide future sequence finishing. Our results demonstrate that eukaryotic genome sequences can be accurately assembled by combining Illumina, 454 and Sanger sequence data.
Figures
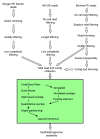
Figure 1
Assembly process overview. Overview of the process for producing de novo assemblies.
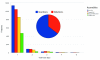
Figure 2
Consensus sequence quality. The proportion of 454 read data within the total read collection affected the number of small insertions and deletions (indels) based on analysis of 7,169 unique EST-to-genome alignments. The relative proportions of insertions (blue) and deletions (orange) in the assembly sequence are shown in the inset pie chart. Assemblies are described in Tables 1 and 2; those including 454 read data were assembled with Forge; the Illumina-only assembly was generated with Velvet.
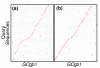
Figure 3
Comparison of Forge Sanger/454/Illumina assemblies against _GC_gb1. Alignments of scaffolds greater that 100 kb - (a) 'Sanger/454/IlluminaDA' (approximately 24 Mb on 80 scaffolds) and (b) 'Sanger/454/IlluminaPA' (approximately 28.7 Mb on 46 scaffolds) - on the y-axis against the manually finished genome sequence (_GC_gb1) on the x-axis.
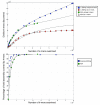
Figure 4
Assessing the discovery of unique read information between the Illumina and 454 platforms. (a) Raw reads were processed into overlapping 28-bp _k_-mers, and any _k_-mer that varied from all other _k_-mers by at least 1 bp was accepted as new sequence information. The analysis was done separately for unique _k_-mers and those that occurred at least twice (2× _k_-mers). (b) MAQ was then used to map these _k_-mers to the reference genome sequence and the rate at which new coverage was generated was plotted against the number of _k_-mers examined.
Similar articles
- De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis.
Nowrousian M, Stajich JE, Chu M, Engh I, Espagne E, Halliday K, Kamerewerd J, Kempken F, Knab B, Kuo HC, Osiewacz HD, Pöggeler S, Read ND, Seiler S, Smith KM, Zickler D, Kück U, Freitag M. Nowrousian M, et al. PLoS Genet. 2010 Apr 8;6(4):e1000891. doi: 10.1371/journal.pgen.1000891. PLoS Genet. 2010. PMID: 20386741 Free PMC article. - A biologist's guide to de novo genome assembly using next-generation sequence data: A test with fungal genomes.
Haridas S, Breuill C, Bohlmann J, Hsiang T. Haridas S, et al. J Microbiol Methods. 2011 Sep;86(3):368-75. doi: 10.1016/j.mimet.2011.06.019. Epub 2011 Jul 3. J Microbiol Methods. 2011. PMID: 21749903 - Fine de novo sequencing of a fungal genome using only SOLiD short read data: verification on Aspergillus oryzae RIB40.
Umemura M, Koyama Y, Takeda I, Hagiwara H, Ikegami T, Koike H, Machida M. Umemura M, et al. PLoS One. 2013 May 7;8(5):e63673. doi: 10.1371/journal.pone.0063673. Print 2013. PLoS One. 2013. PMID: 23667655 Free PMC article. - De novo assembly of short sequence reads.
Paszkiewicz K, Studholme DJ. Paszkiewicz K, et al. Brief Bioinform. 2010 Sep;11(5):457-72. doi: 10.1093/bib/bbq020. Epub 2010 Aug 19. Brief Bioinform. 2010. PMID: 20724458 Review. - Discovering the hidden function in fungal genomes.
Gervais NC, Shapiro RS. Gervais NC, et al. Nat Commun. 2024 Sep 19;15(1):8219. doi: 10.1038/s41467-024-52568-z. Nat Commun. 2024. PMID: 39300175 Free PMC article. Review.
Cited by
- Sequencing of the Dutch elm disease fungus genome using the Roche/454 GS-FLX Titanium System in a comparison of multiple genomics core facilities.
Forgetta V, Leveque G, Dias J, Grove D, Lyons R Jr, Genik S, Wright C, Singh S, Peterson N, Zianni M, Kieleczawa J, Steen R, Perera A, Bintzler D, Adams S, Hintz W, Jacobi V, Bernier L, Levesque R, Dewar K. Forgetta V, et al. J Biomol Tech. 2013 Apr;24(1):39-49. doi: 10.7171/jbt.12-2401-005. J Biomol Tech. 2013. PMID: 23542132 Free PMC article. - RNA-seq analyses of blood-induced changes in gene expression in the mosquito vector species, Aedes aegypti.
Bonizzoni M, Dunn WA, Campbell CL, Olson KE, Dimon MT, Marinotti O, James AA. Bonizzoni M, et al. BMC Genomics. 2011 Jan 28;12:82. doi: 10.1186/1471-2164-12-82. BMC Genomics. 2011. PMID: 21276245 Free PMC article. - An NGS Workflow Blueprint for DNA Sequencing Data and Its Application in Individualized Molecular Oncology.
Li J, Batcha AM, Grüning B, Mansmann UR. Li J, et al. Cancer Inform. 2016 Apr 10;14(Suppl 5):87-107. doi: 10.4137/CIN.S30793. eCollection 2015. Cancer Inform. 2016. PMID: 27081306 Free PMC article. Review. - Methods to improve the accuracy of next-generation sequencing.
Cheng C, Fei Z, Xiao P. Cheng C, et al. Front Bioeng Biotechnol. 2023 Jan 20;11:982111. doi: 10.3389/fbioe.2023.982111. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 36741756 Free PMC article. Review. - Construction of a public CHO cell line transcript database using versatile bioinformatics analysis pipelines.
Rupp O, Becker J, Brinkrolf K, Timmermann C, Borth N, Pühler A, Noll T, Goesmann A. Rupp O, et al. PLoS One. 2014 Jan 10;9(1):e85568. doi: 10.1371/journal.pone.0085568. eCollection 2014. PLoS One. 2014. PMID: 24427317 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous