A robust two-dimensional separation for top-down tandem mass spectrometry of the low-mass proteome - PubMed (original) (raw)
. 2009 Dec;20(12):2183-91.
doi: 10.1016/j.jasms.2009.08.001. Epub 2009 Aug 12.
John F Kellie, John C Tran, Jeremiah D Tipton, Adam D Catherman, Haylee M Thomas, Dorothy R Ahlf, Kenneth R Durbin, Adaikkalam Vellaichamy, Ioanna Ntai, Alan G Marshall, Neil L Kelleher
Affiliations
- PMID: 19747844
- PMCID: PMC2830800
- DOI: 10.1016/j.jasms.2009.08.001
A robust two-dimensional separation for top-down tandem mass spectrometry of the low-mass proteome
Ji Eun Lee et al. J Am Soc Mass Spectrom. 2009 Dec.
Abstract
For fractionation of intact proteins by molecular weight (MW), a sharply improved two-dimensional (2D) separation is presented to drive reproducible and robust fractionation before top-down mass spectrometry of complex mixtures. The "GELFrEE" (i.e., gel-eluted liquid fraction entrapment electrophoresis) approach is implemented by use of Tris-glycine and Tris-tricine gel systems applied to human cytosolic and nuclear extracts from HeLa S3 cells, to achieve a MW-based fractionation of proteins from 5 to >100 kDa in 1 h. For top-down tandem mass spectroscopy (MS/MS) of the low-mass proteome (5-25 kDa), between 5 and 8 gel-elution (GE) fractions are sampled by nanocapillary-LC-MS/MS with 12 or 14.5 tesla Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometers. Single injections give about 40 detectable proteins, about half of which yield automated ProSight identifications. Reproducibility metrics of the system are presented, along with comparative analysis of protein targets in mitotic versus asynchronous cells. We forward this basic 2D approach to facilitate wider implementation of top-down mass spectrometry and a variety of other protein separation and/or characterization approaches.
Figures
Figure 1
Workflow for top-down proteomics. Total protein content from HeLa-S3 nuclei or cytosol is quantified and loaded onto a gel-elution (GE) column. The GE device separates the protein samples according to molecular weight (MW). Proteins of increasing MW elute into solution-phase fractions, which can be visualized on a slab gel (top right). The solution-phase fractions are cleaned up to remove SDS before injection onto a nano-liquid chromatography (LC) column for tandem mass spectrometric (MS/MS) analysis with an LTQ-FT at either 12 or 14.5 T. LC-MS/MS files are processed with ProSightPC 2.0, a software suite tailored for top-down analysis in a high-throughput setting.
Figure 2
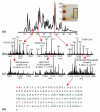
Examples selected from an LC-MS/MS injection of fraction #3 from a Tris-glycine GE run. (a) A base-peak chromatogram is shown, with (b) charge state distributions from selected retention times. (c) Abundant charge states (above the arrows) were targeted for fragmentation. Fragmentation mass spectra for each protein are shown along with the corresponding identifications and E-values. A fragmentation map (d) results from the matching fragment ions of nucleoside diphosphate kinase B. The protein is N-terminally acetylated.
Figure 3
Analytical slab gel and heat map showing masses detected in GE fractions. The silver-stained slab gel (top) of solution-phase fractions shows typical GE separation (Tris-tricine buffer system). After LC-MS of the GE fractions, the detected masses for each fraction are plotted as a function of LC retention time (bottom). The MS relative abundance is designated with the scale on the right; the inset illustrates the enlarged region of heat map of GE fraction 6. General correlation can be seen between detected masses and abundances from the MS and the intensities of protein bands detected on the slab gel.
Figure 4
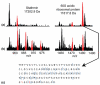
Differences in phosphorylation for proteins identified in asynchronous and M-phase arrested HeLa cells. (a) Asynchronous HeLa proteins are identified with the phosphorylations designated in red. (b) M-phase arrested HeLa proteins are identified, again with phosphorylations designated in red. Dynamic changes in phosphorylation between asynchronous and M-phase arrested HeLa cells were clearly observed. (c) A fragment map for the doubly phosphorylated 60S acidic ribosomal protein (phosphorylation sites in red text).
Similar articles
- Low-Molecular-Weight Plasma Proteome Analysis Using Top-Down Mass Spectrometry.
Cheon DH, Yang EG, Lee C, Lee JE. Cheon DH, et al. Methods Mol Biol. 2017;1619:103-117. doi: 10.1007/978-1-4939-7057-5_8. Methods Mol Biol. 2017. PMID: 28674880 - A Top-Down Proteomics Platform Coupling Serial Size Exclusion Chromatography and Fourier Transform Ion Cyclotron Resonance Mass Spectrometry.
Tucholski T, Knott SJ, Chen B, Pistono P, Lin Z, Ge Y. Tucholski T, et al. Anal Chem. 2019 Mar 19;91(6):3835-3844. doi: 10.1021/acs.analchem.8b04082. Epub 2019 Feb 25. Anal Chem. 2019. PMID: 30758949 Free PMC article. - Identification and Characterization of Human Proteoforms by Top-Down LC-21 Tesla FT-ICR Mass Spectrometry.
Anderson LC, DeHart CJ, Kaiser NK, Fellers RT, Smith DF, Greer JB, LeDuc RD, Blakney GT, Thomas PM, Kelleher NL, Hendrickson CL. Anderson LC, et al. J Proteome Res. 2017 Feb 3;16(2):1087-1096. doi: 10.1021/acs.jproteome.6b00696. Epub 2016 Dec 12. J Proteome Res. 2017. PMID: 27936753 Free PMC article. - Screening of synthetic PDE-5 inhibitors and their analogues as adulterants: analytical techniques and challenges.
Patel DN, Li L, Kee CL, Ge X, Low MY, Koh HL. Patel DN, et al. J Pharm Biomed Anal. 2014 Jan;87:176-90. doi: 10.1016/j.jpba.2013.04.037. Epub 2013 May 6. J Pharm Biomed Anal. 2014. PMID: 23721687 Review. - Prefractionation of protein samples for proteome analysis using reversed-phase high-performance liquid chromatography.
Badock V, Steinhusen U, Bommert K, Otto A. Badock V, et al. Electrophoresis. 2001 Aug;22(14):2856-64. doi: 10.1002/1522-2683(200108)22:14<2856::AID-ELPS2856>3.0.CO;2-U. Electrophoresis. 2001. PMID: 11565780 Review.
Cited by
- Top down proteomics of human membrane proteins from enriched mitochondrial fractions.
Catherman AD, Li M, Tran JC, Durbin KR, Compton PD, Early BP, Thomas PM, Kelleher NL. Catherman AD, et al. Anal Chem. 2013 Feb 5;85(3):1880-8. doi: 10.1021/ac3031527. Epub 2013 Jan 23. Anal Chem. 2013. PMID: 23305238 Free PMC article. - High-throughput middle-down analysis using an orbitrap.
Cannon J, Lohnes K, Wynne C, Wang Y, Edwards N, Fenselau C. Cannon J, et al. J Proteome Res. 2010 Aug 6;9(8):3886-90. doi: 10.1021/pr1000994. J Proteome Res. 2010. PMID: 20557100 Free PMC article. - Post-translational modifications of integral membrane proteins resolved by top-down Fourier transform mass spectrometry with collisionally activated dissociation.
Ryan CM, Souda P, Bassilian S, Ujwal R, Zhang J, Abramson J, Ping P, Durazo A, Bowie JU, Hasan SS, Baniulis D, Cramer WA, Faull KF, Whitelegge JP. Ryan CM, et al. Mol Cell Proteomics. 2010 May;9(5):791-803. doi: 10.1074/mcp.M900516-MCP200. Epub 2010 Jan 21. Mol Cell Proteomics. 2010. PMID: 20093275 Free PMC article. - Quantitative fluorescence labeling of aldehyde-tagged proteins for single-molecule imaging.
Shi X, Jung Y, Lin LJ, Liu C, Wu C, Cann IK, Ha T. Shi X, et al. Nat Methods. 2012 Apr 1;9(5):499-503. doi: 10.1038/nmeth.1954. Nat Methods. 2012. PMID: 22466795 Free PMC article. - Analysis of protein isoforms: can we do it better?
Stastna M, Van Eyk JE. Stastna M, et al. Proteomics. 2012 Oct;12(19-20):2937-48. doi: 10.1002/pmic.201200161. Epub 2012 Sep 19. Proteomics. 2012. PMID: 22888084 Free PMC article. Review.
References
- Kelleher NL, Costello CA, Begley TP, McLafferty FW. Thiaminase-I (42-kDa) Heterogeneity, Sequence Refinement, and Active-Site Location from High-Resolution Tandem Mass-Spectrometry. J. Am. Soc. Mass Spectrom. 1995;6:981–984. - PubMed
- Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. Top Down versus Bottom Up Protein Characterization by Tandem High-Resolution Mass Spectrometry. J. Am. Chem. Soc. 1999;121:806–812.
- McLafferty FW. High-Resolution Tandem FT Mass-Spectrometry above 10-kDa. Acc. Chem. Res. 1994;27:379–386.
- Chong BE, Yan F, Lubman DM, Miller FR. Chromatofocusing Nonporous Reversed-Phase High-Performance Liquid Chromatography/Electrospray Ionization Time-of-Flight Mass Spectrometry of Proteins from Human Breast Cancer Whole Cell Lysates: A Novel Two-Dimensional Liquid Chromatography/Mass Spectrometry Method. Rapid Commun. Mass Spectrom. 2001;15:291–296. - PubMed
- Lubman DM, Kachman MT, Wang HX, Gong SY, Yan F, Hamler RL, O’Neil KA, Zhu K, Buchanan NS, Barder TJ. Two-Dimensional Liquid Separations—Mass Mapping of Proteins from Human Cancer Cell Lysates. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2002;782:183–196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM067193-07/GM/NIGMS NIH HHS/United States
- P30 DA018310/DA/NIDA NIH HHS/United States
- R01 GM067193/GM/NIGMS NIH HHS/United States
- GM-067193-07/GM/NIGMS NIH HHS/United States
- P30 DA-018310-06/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials