An intersubunit signaling network coordinates ATP hydrolysis by m-AAA proteases - PubMed (original) (raw)
An intersubunit signaling network coordinates ATP hydrolysis by m-AAA proteases
Steffen Augustin et al. Mol Cell. 2009.
Abstract
Ring-shaped AAA+ ATPases control a variety of cellular processes by substrate unfolding and remodeling of macromolecular structures. However, how ATP hydrolysis within AAA+ rings is regulated and coupled to mechanical work is poorly understood. Here we demonstrate coordinated ATP hydrolysis within m-AAA protease ring complexes, conserved AAA+ machines in the inner membrane of mitochondria. ATP binding to one AAA subunit inhibits ATP hydrolysis by the neighboring subunit, leading to coordinated rather than stochastic ATP hydrolysis within the AAA ring. Unbiased genetic screens define an intersubunit signaling pathway involving conserved AAA motifs and reveal an intimate coupling of ATPase activities to central AAA pore loops. Coordinated ATP hydrolysis between adjacent subunits is required for membrane dislocation of substrates, but not for substrate processing. These findings provide insight into how AAA+ proteins convert energy derived from ATP hydrolysis into mechanical work.
Figures
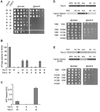
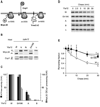
Figure 1. Coordinated ATP hydrolysis within yeast _m_-AAA protease complexes
(A) Growth of Δ_yta10_Δ_yta12_ cells on fermentable (glucose) and non-fermentable (glycerol) carbon sources upon expression of wild type and mutant variants of Yta10 and Yta12. W/W, Yta10-6His/Yta12; W/A, Yta10-6His/Yta12K394A; A/W, Yta10K334A-6His/Yta12; A/A, Yta10K334A-6His/Yta12K394A; W/B, Yta10-6His/Yta12E448Q; B/W, Yta10E388Q-6His/Yta12; B/B, Yta10E388Q-6His/Yta12E448Q; −/−, vector control. (B) ATPase activities of wild type and mutant m_-AAA proteases in vitro. W/−, Yta10-6His/empty vector; −/W, empty vector/Yta12-6His. (C) Detection of trapped ATP co-purifying with m_-AAA protease variants by a luciferase-based assay. Data represent the mean +/− standard deviation of three independent experiments. AU, arbitrary units. (D) Upper panel, schematic representation of Yta10 and an Yta12-derived chimeric subunit harboring the AAA domain of Yta10 (H10). Lower panel, growth of Δ_yta10_Δ_yta12 cells expressing Yta10 (10W) and Yta12 (12W) or variants thereof as indicated. MTS, mitochondrial targeting sequence; ND, N-terminal domain; AAA, AAA domain; PD, proteolytic domain; CH, C-terminal helices. (E) Upper panel, schematic representation of Yta12 and an Yta10-derived chimeric subunit harboring the AAA domain of Yta12 (H12). Lower panel, growth of Δ_yta10_Δ_yta12 cells expressing Yta12 (12W) and Yta10 (10W) or variants thereof as indicated.
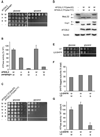
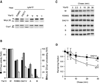
Figure 2. Dominant negative effects of mutations in the Walker B motif on homo- and hetero-oligomeric human _m_-AAA proteases
(A) Growth of Δ_yta10_Δ_yta12_ cells on fermentable (glucose) and non-fermentable (glycerol) carbon sources upon co-expression of wild type paraplegin with AFG3L2 or its mutant variants. W/W, AFG3L2/paraplegin-6His; A/W, AFG3L2K354A/paraplegin-6His; B/W, AFG3L2E408Q/paraplegin-6His −/−, vector control (YCplac22/YEplac181). (B) In vitro ATPase activities of hetero-oligomeric human m_-AAA proteases harboring mutations in Walker A or B motifs. Activities were normalized to the number of assembled complexes. W/A, AFG3L2/parapleginK355A-6His; W/B, AFG3L2/ parapleginE409Q-6His. (C) Growth of Δ_yta10_Δ_yta12 cells upon co-expression of wild type and mutant variants of AFG3L2 harboring mutations within Walker A or B motifs. W/W, AFG3L2/AFG3L2; W/A, AFG3L2/AFG3L2K354A; W/B, AFG3L2/AFG3L2E408Q; A/A, AFG3L2K354A/AFG3L2K354A; B/B, AFG3L2 E408Q/AFG3L2E408Q; −/−, vector control. (D) Processing of MrpL32 and Ccp1 in Δ_yta10_Δ_yta12_ cells expressing AFG3L2 and mutant variants thereof. Total proteins were extracted by alkaline lysis following separation by SDS-PAGE and immune blotting using antisera directed against Ccp1, MrpL32, AFG3L2 and Tom40. p, precursor; m, mature. (E) Respiratory competence of Δ_yta10_Δ_yta12_ cells expressing AFG3L2 (L2) or DHFR-tagged AFG3L2 (L2-DHFR). (F) Subunit stoichiometry in homo-oligomeric human _m_-AAA proteases composed of DHFR- and his-tagged subunits harboring mutations when indicated. After SDS-PAGE and coomassie staining analysis of purified complexes, bands were densitometorically quantified. The proportion of DHFR-tagged subunits is shown. Since _m_-AAA proteases were purified by metal chelating chromatography from the pool of stochastically formed complexes, his-tagged subunits are overrepresented in the purified fraction. (G) In vitro ATPase activities of human AFG3L2 complexes composed of L2-DHFR and AFG3L2 subunits carrying mutations in Walker A or B motifs when indicated. Activities were normalized to the number of assembled wild type subunits in the complexes. Data in (B) and (G) represent the mean +/− standard deviation of three independent experiments.
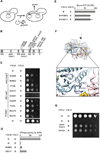
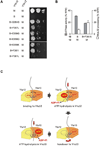
Figure 3. Identification of amino acid residues in Yta10 involved in intersubunit signaling
(A) Screen for suppressor mutations in Yta10 allowing respiratory growth in the presence of a mutation in the Walker B motif of Yta12. (B) Suppressor mutations in Yta10. Conserved motifs are indicated by grey bars. The number of identified clones harboring the specific mutation is given in brackets. WA, Walker A motif; WB, Walker B motif; S1, sensor-1 motif; R, arginine finger; ISS, intersubunit signaling motif. (C) Respiratory growth of Δ_yta10_Δ_yta12_ cells co-expressing dominant negative Yta12E448Q (B) and suppressing alleles of Yta10. (D) ATPase activities of mutant _m_-AAA protease complexes harboring Yta12E448Q and suppressor alleles of Yta10. (E) ATP binding to mutant _m_-AAA protease complexes composed of Yta12E448Q subunits and Yta10E388Q subunits harboring suppressor alleles. ATP co-purifying with _m_-AAA protease variants was detected using a luciferase-based assay. Data represent the mean +/− standard deviation of three independent experiments. (F) Localization of suppressors in Yta10 based on an atomic model of the yeast _m_-AAA protease hexamer. Yta10 is shown in cyan and Yta12 in pink. For clarity, only one pair of Yta10 and Yta12 is colored. Identified suppressor mutations and conserved glutamates in the Walker B motifs are shown as stick model. Bound nucleotide and magnesium ions are shown as CPK model and colored gold and green, respectively. The position of the 6-fold symmetry axis is indicated. The inset depicts a close-up view of the Yta10/Yta12 interface and shows the position of pore loop-2 of Yta10. (G) Deletion of the pore loop-2 or a mutation of the arginine residue R450 in Yta10 also suppresses the respiratory incompetence caused by the mutation in the Walker B motif of Yta12.
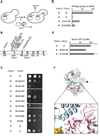
Figure 4. Identification of amino acid residues in Yta12 involved in intersubunit signaling
(A) Screen for intragenic suppressor mutations in Yta12 allowing respiratory growth in the presence of mutations in the Walker B motif of Yta12. (B) Intragenic suppressor mutations in Yta12. Conserved motifs are indicated as in Fig. 3B. (C) Respiratory competence of Δ_yta10_Δ_yta12_ cells which co-express wild type Yta10 (W) and Yta12E448Q (B) harboring suppressor mutations. (D) ATPase activities of mutant _m_-AAA protease complexes composed of Yta10 subunits and Yta12E448Q variants harboring intragenic suppressor mutations. (E) ATP binding to mutant _m_-AAA protease complexes composed of Yta10E388Q subunits and Yta12E448Q subunits harboring suppressor alleles. ATP co-purifying with _m_-AAA protease variants was detected using a luciferase-based assay. Data represent the mean +/− standard deviation of three independent experiments. (F) Localization of suppressors in Yta12 based on an atomic model of the yeast _m_-AAA protease hexamer. Yta10 and Yta12 subunits and other key features are shown as described in Fig. 3F. The inset depicts a close-up view of the Yta10/Yta12 interface and shows the position of pore loop-1 and -2 of Yta12.
Figure 5. Intersubunit signaling within Yta12 and substrate handling by the _m_-AAA protease
(A) Schematic representation of proteolytic substrates of the _m_-AAA protease. Transmembrane segments are shown as black boxes, folded domains as circles. Ccp1 but not MrpL32 processing depends on ATP-dependent membrane dislocation driven by the m_-AAA protease (Tatsuta et al., 2007). Yme2ΔC is extracted from the membrane and degraded processively (Leonhard et al., 2000). (B) Processing of MrpL32 and Ccp1 in Δ_yta12 cells (Δ) expressing Yta12 or Yta12 variants harboring mutations in the Walker A- (K394A) or the Walker B- (E448Q) motif or a suppressor mutation preceding the pore loop-1 (E416K). p, precursor form of MrpL32 or Ccp1; m, mature form of MrpL32 or Ccp1; asterisk, intermediate form of MrpL32. (C) ATPase activities of purified m_-AAA protease variants containing mutant Yta12 subunits in vitro and their processing activity (Ccp1, MrpL32) in vivo. (D) Stability of Yme2ΔC in mitochondria. 35S-labeled Yme2ΔC was imported into mitochondria harbouring Yta10 and variants of Yta12 as indicated. After removal of non-imported precursor proteins by trypsin, samples were incubated at 37°C for the time indicated to allow proteolysis to occur. (E) 35S-labeled mature Yme2ΔC was quantified at different time points by phosphoimaging. Newly imported Yme2ΔC present in mitochondria before the temperature shift to 37°C was set to 1. Data represent the mean +/− standard deviation of three independent experiments. Open circles, WT (W); open diamonds, Δ_yta12 (Δ); closed squares, Yta12E416K (E416K).
Figure 6. Intersubunit signaling in Yta10 and substrate handling by the _m_-AAA protease
(A) Processing of MrpL32 and Ccp1 in Δ_yta10_ cells expressing Yta10 or Yta10 variants harboring mutations in the Walker A- (K334A) or Walker B- (E388Q) motif or mutations in pore loop-2 (R396G) or the ISS-motif (D421V) which prevent intersubunit signaling. Membrane fractions (MrpL32) or total cell lysates (Ccp1) were analyzed by SDS-PAGE and immune blotting using antisera directed against Ccp1 and MrpL32. p, precursor form of MrpL32 or Ccp1; m, mature form of MrpL32 or Ccp1; asterisk, intermediate form of MrpL32. (B) ATPase activities of purified m_-AAA proteases containing mutant Yta10 subunits in vitro and their processing activity (Ccp1, MrpL32) in vivo. (C) Stability of Yme2ΔC in mitochondria. 35S-labeled Yme2ΔC was imported into mitochondria harbouring Yta12 and variants of Yta10 as indicated and the stability was assessed as in Fig. 5D. (D) 35S-labeled mature Yme2ΔC was quantified at different time points by phosphoimaging as in Fig. 5E. Data represent the mean +/− standard deviation of three independent experiments. Open circles, WT (W); open diamonds, Δ_yta12 (Δ); closed squares, Yta12D421V (D421V).
Figure 7. Role of pore loop-1 for coupling of ATPase and proteolytic activities of _m_-AAA proteases
(A) Respiratory growth of Δ_yta10_Δ_yta12_ cells expressing Yta12 (W) and Yta10E388Q (B) variants harboring intragenic suppressors. Intragenic suppressor mutations in Yta10E388Q were identified analogous to the gain-of-function screen described in Fig. 4A. (B) Correlation of MrpL32 processing in vivo with the ATPase activity of a mutant _m_-AAA protease composed of Yta12 and Yta10E388Q (B) containing a mutation in the pore loop-1 (F361A) when indicated. Data represent the mean +/− standard deviation of three independent experiments. (C) Coordinated ATP hydrolysis ensures substrate handover between adjacent subunits. See text for details. A heterodimer of Yta10 and Yta12 subunits as the minimal functional unit of the yeast _m_-AAA protease is shown.
Comment in
- The alternating power stroke of a 6-cylinder AAA protease chaperone engine.
Kress W, Weber-Ban E. Kress W, et al. Mol Cell. 2009 Sep 11;35(5):545-7. doi: 10.1016/j.molcel.2009.08.013. Mol Cell. 2009. PMID: 19748349
Similar articles
- The alternating power stroke of a 6-cylinder AAA protease chaperone engine.
Kress W, Weber-Ban E. Kress W, et al. Mol Cell. 2009 Sep 11;35(5):545-7. doi: 10.1016/j.molcel.2009.08.013. Mol Cell. 2009. PMID: 19748349 - Substrate specific consequences of central pore mutations in the i-AAA protease Yme1 on substrate engagement.
Graef M, Langer T. Graef M, et al. J Struct Biol. 2006 Oct;156(1):101-8. doi: 10.1016/j.jsb.2006.01.009. Epub 2006 Feb 21. J Struct Biol. 2006. PMID: 16527490 - Substrate recognition by AAA+ ATPases: distinct substrate binding modes in ATP-dependent protease Yme1 of the mitochondrial intermembrane space.
Graef M, Seewald G, Langer T. Graef M, et al. Mol Cell Biol. 2007 Apr;27(7):2476-85. doi: 10.1128/MCB.01721-06. Epub 2007 Jan 29. Mol Cell Biol. 2007. PMID: 17261594 Free PMC article. - Stairway to translocation: AAA+ motor structures reveal the mechanisms of ATP-dependent substrate translocation.
Gates SN, Martin A. Gates SN, et al. Protein Sci. 2020 Feb;29(2):407-419. doi: 10.1002/pro.3743. Epub 2019 Oct 17. Protein Sci. 2020. PMID: 31599052 Free PMC article. Review. - ATP-dependent proteases controlling mitochondrial function in the yeast Saccharomyces cerevisiae.
Van Dyck L, Langer T. Van Dyck L, et al. Cell Mol Life Sci. 1999 Nov 30;56(9-10):825-42. doi: 10.1007/s000180050029. Cell Mol Life Sci. 1999. PMID: 11212342 Free PMC article. Review.
Cited by
- Alternative splicing of Spg7, a gene involved in hereditary spastic paraplegia, encodes a variant of paraplegin targeted to the endoplasmic reticulum.
Mancuso G, Barth E, Crivello P, Rugarli EI. Mancuso G, et al. PLoS One. 2012;7(5):e36337. doi: 10.1371/journal.pone.0036337. Epub 2012 May 1. PLoS One. 2012. PMID: 22563492 Free PMC article. - Mutations in the mitochondrial protease gene AFG3L2 cause dominant hereditary ataxia SCA28.
Di Bella D, Lazzaro F, Brusco A, Plumari M, Battaglia G, Pastore A, Finardi A, Cagnoli C, Tempia F, Frontali M, Veneziano L, Sacco T, Boda E, Brussino A, Bonn F, Castellotti B, Baratta S, Mariotti C, Gellera C, Fracasso V, Magri S, Langer T, Plevani P, Di Donato S, Muzi-Falconi M, Taroni F. Di Bella D, et al. Nat Genet. 2010 Apr;42(4):313-21. doi: 10.1038/ng.544. Epub 2010 Mar 7. Nat Genet. 2010. PMID: 20208537 - Unique double-ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo-electron microscopy.
Blok NB, Tan D, Wang RY, Penczek PA, Baker D, DiMaio F, Rapoport TA, Walz T. Blok NB, et al. Proc Natl Acad Sci U S A. 2015 Jul 28;112(30):E4017-25. doi: 10.1073/pnas.1500257112. Epub 2015 Jul 13. Proc Natl Acad Sci U S A. 2015. PMID: 26170309 Free PMC article. - Structural basis for intersubunit signaling in a protein disaggregating machine.
Biter AB, Lee S, Sung N, Tsai FT. Biter AB, et al. Proc Natl Acad Sci U S A. 2012 Jul 31;109(31):12515-20. doi: 10.1073/pnas.1207040109. Epub 2012 Jul 16. Proc Natl Acad Sci U S A. 2012. PMID: 22802670 Free PMC article. - Regulated protein turnover: snapshots of the proteasome in action.
Bhattacharyya S, Yu H, Mim C, Matouschek A. Bhattacharyya S, et al. Nat Rev Mol Cell Biol. 2014 Feb;15(2):122-33. doi: 10.1038/nrm3741. Nat Rev Mol Cell Biol. 2014. PMID: 24452470 Free PMC article. Review.
References
- Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10-12-complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. - PubMed
- Bolon DN, Grant RA, Baker TA, Sauer RT. Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol. Cell. 2004;16:343–350. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases