Canalization of gene expression in the Drosophila blastoderm by gap gene cross regulation - PubMed (original) (raw)
Canalization of gene expression in the Drosophila blastoderm by gap gene cross regulation
Manu et al. PLoS Biol. 2009 Mar.
Abstract
Developing embryos exhibit a robust capability to reduce phenotypic variations that occur naturally or as a result of experimental manipulation. This reduction in variation occurs by an epigenetic mechanism called canalization, a phenomenon which has resisted understanding because of a lack of necessary molecular data and of appropriate gene regulation models. In recent years, quantitative gene expression data have become available for the segment determination process in the Drosophila blastoderm, revealing a specific instance of canalization. These data show that the variation of the zygotic segmentation gene expression patterns is markedly reduced compared to earlier levels by the time gastrulation begins, and this variation is significantly lower than the variation of the maternal protein gradient Bicoid. We used a predictive dynamical model of gene regulation to study the effect of Bicoid variation on the downstream gap genes. The model correctly predicts the reduced variation of the gap gene expression patterns and allows the characterization of the canalizing mechanism. We show that the canalization is the result of specific regulatory interactions among the zygotic gap genes. We demonstrate the validity of this explanation by showing that variation is increased in embryos mutant for two gap genes, Krüppel and knirps, disproving competing proposals that canalization is due to an undiscovered morphogen, or that it does not take place at all. In an accompanying article in PLoS Computational Biology (doi:10.1371/journal.pcbi.1000303), we show that cross regulation between the gap genes causes their expression to approach dynamical attractors, reducing initial variation and providing a robust output. These results demonstrate that the Bicoid gradient is not sufficient to produce gap gene borders having the low variance observed, and instead this low variance is generated by gap gene cross regulation. More generally, we show that the complex multigenic phenomenon of canalization can be understood at a quantitative and predictive level by the application of a precise dynamical model.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
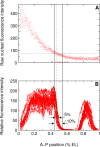
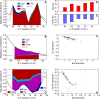
Figure 1. Comparison of In Vivo Bcd Variability with Hb Variability
(A) Fifteen Bcd-GFP concentration profiles replotted from the data of Gregor and colleagues (Figure 5A in [16]). (B) Eighteen Hb profiles 3 min before gastrulation (time class T8, see Table 1) from FlyEx [6]. The dotted vertical line is at the average position of hb border. The dotted horizontal line passes through the point where the dotted vertical line crosses the middle of the Bcd scatter. The solid black lines delineate the horizontal spread of Bcd. The red tick marks have been placed uniformly using the graphics program Xfig.
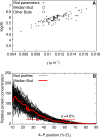
Figure 2. Selection of a Representative Bcd Profile
(A) Scatter plot of log amplitude (log A) and slope (γ) of 88 Bcd profiles from cycle 13 embryos. The boxed profile and the circled profiles were investigated further; unless explicitly mentioned, the analysis in this paper uses the boxed (median) profile. (B) An overlay of all 88 Bcd profiles used in the simulations. The median profile is highlighted in red. The threshold concentration at which the Hb border forms in the gene circuit was determined from the median profile. The positions at which these 88 profiles cross that threshold has a range of 20.6% EL, and a standard deviation (γ) of 4.6% EL.
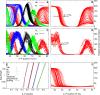
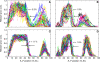
Figure 3. Gene Circuits Correctly Predict the Variation of Six Gap Gene Domain Borders
(A) Modeled expression patterns of hb, Kr, gt, and kni in time class T8 (see Table 1) in simulations of the gene circuit using Bcd profiles from 88 individual embryos; compare with (C). (B) hb patterns produced by the model are shown separately. The position of the posterior border of the anterior hb domain has σHb= 1.3% EL and ρHb = 5.6% EL. (C) Gap gene expression data from 83 time class T8 wild-type embryos. There are 15 expression profiles for hb, 30 for Kr, 18 for gt, and 15 for kni. (D) σHb = 1.0% EL and ρHb = 3.6% EL in data. (E) Simulation of size variation. The egg length was varied from 0.9 to 1.1 to simulate a 20% range of egg lengths. The _x_-axis is the absolute position (relative position × length) of the gap gene borders. The border positions of the six gap gene domain borders that have low variance under Bcd variation (A) are plotted as points. The solid black lines show idealized proportional scaling of border positions with egg length. (F) In silico control experiment of simulating Bcd variation with gap gene cross regulation of hb turned off. The Bcd profiles used were the same as the ones used to produce (A) and (B). Fourteen borders are located anterior to 35% EL and are not visible in the region shown. σHb = 4.3% EL and ρHb = 19.6% EL.
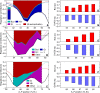
Figure 4. Regulatory Analysis of the Posterior Borders of the Anterior hb, the Central Kr, and the Posterior kni Domains
(A–C) Analysis of the chosen gene circuit (see Results section “Simulation of Bcd Variation and Size Variation”) with the median Bcd profile (Figure 2B). Dashed vertical lines demarcate a border, and correspond to positions where the expression is at 90% maximum and at 10% maximum. The solid black line is the spatial derivative of the total regulatory input to the gene ua (see Section S3 in Protocol S1). The area above the black line is the total change in total regulatory input ua that causes the border to form. The colored areas correspond to the contributions to the change in total input by different regulators of the gene. The regulatory inputs that cannot set a border are shown in red (see Section S3 in Protocol S1 for details), and are not included in the analysis. A regulatory contribution can be from an activator or a repressor depending on the sign of the regulatory parameter (see Table S1 in Protocol S1 for values). (A) The hb border forms because of the regulatory contributions of Bcd activation, Kr repression, and Kni repression. The contribution of Hb autoactivation, shown in red, does not set the border but merely sharpens it [9,36]. The colored bar inset shows the relative contributions of Bcd activation, Kr repression, and Kni repression to hb, showing that the repressive contribution is significant compared to the activating one. (B) The Kr border is set by Bcd activation and repression from Kni and Gt. (C) The kni border is set by repression from Gt and Hb. (D–I) The effects of simulated Bcd variation on the regulation of gap gene borders. (D–F) The average Bcd activation at a border in 88 simulations (with different Bcd profiles) pooled into 1% EL bins according to border position. (G–I) The average repression levels at a border in 88 simulations pooled into 1% EL bins according to border position. The repressors were identified in the regulatory analysis (A–C). The _x-_axis shows bin position. The analysis was performed on model output in time class T8.
Figure 5. Regulatory Analysis of the Anterior Border of the Posterior kni Domain and Both Borders of the Posterior gt Domain
(A–C) Analysis of the chosen gene circuit (see Results section “Simulation of Bcd Variation and Size Variation”) with the median Bcd profile (Figure 2B). The graphical regulatory analysis is described in the caption of Figure 4. (A) The kni border forms because of the regulatory contributions of Cad activation, Kr repression, and Hb repression. (B) The anterior gt border is set by Cad activation and repression from Kr. (C) The posterior gt border is set by Bcd activation and repression from Hb and Tll. (D–G) The effects of simulated Bcd variation on the regulation of gap gene borders. (D) The average Bcd activation at the kni border in 88 simulations (with different Bcd profiles) pooled into 1% EL bins according to border position. (E) The average repression levels at the kni border in 88 simulations pooled into 1% EL bins according to border position. (F) Scatter plot of the Bcd activation and Kr repression at the anterior gt border in each of the 88 simulations. (G) Scatter plot of the Bcd activation and Hb/Tll repression at the posterior gt border in each of the 88 simulations. The repressors were identified in the regulatory analysis (A–C). The _x-_axis shows bin position. The analysis was performed on model output in time class T8.
Figure 6. The Control of Positional Variation of the Posterior Border of the Anterior hb Domain by Kr and Kni
hb (A) and gt (B) expression profiles in 28 Kr;kni double mutant embryos from time classes T4–T7 (Table 1); σHb = 2.2% EL (standard deviation) and ρHb = 9.3% EL (range). The standard deviation of the posterior border of the third gt domain is σGt = 1.9% EL; ρGt = 8.9% EL. hb (C) and gt (D) expression profiles in wild-type embryos of the same age as in (A) and (B). σHb = 1.1% EL and ρHb = 5.6% EL (68 embryos). σGt = 1.2% EL and ρGt = 5.6% EL (92 embryos). Expression patterns are shown normalized against the maximum level (see Methods) in the anterior hb (A and C) or the anterior gt (B and D) domains.
Similar articles
- Mechanisms of gap gene expression canalization in the Drosophila blastoderm.
Gursky VV, Panok L, Myasnikova EM, Manu, Samsonova MG, Reinitz J, Samsonov AM. Gursky VV, et al. BMC Syst Biol. 2011;5:118. doi: 10.1186/1752-0509-5-118. Epub 2011 Jul 28. BMC Syst Biol. 2011. PMID: 21794172 Free PMC article. - Canalization of gene expression and domain shifts in the Drosophila blastoderm by dynamical attractors.
Manu, Surkova S, Spirov AV, Gursky VV, Janssens H, Kim AR, Radulescu O, Vanario-Alonso CE, Sharp DH, Samsonova M, Reinitz J. Manu, et al. PLoS Comput Biol. 2009 Mar;5(3):e1000303. doi: 10.1371/journal.pcbi.1000303. Epub 2009 Mar 13. PLoS Comput Biol. 2009. PMID: 19282965 Free PMC article. - A precise Bicoid gradient is nonessential during cycles 11-13 for precise patterning in the Drosophila blastoderm.
Lucchetta EM, Vincent ME, Ismagilov RF. Lucchetta EM, et al. PLoS One. 2008;3(11):e3651. doi: 10.1371/journal.pone.0003651. Epub 2008 Nov 7. PLoS One. 2008. PMID: 18989373 Free PMC article. - The mother-to-child transition.
Tadros W, Westwood JT, Lipshitz HD. Tadros W, et al. Dev Cell. 2007 Jun;12(6):847-9. doi: 10.1016/j.devcel.2007.05.009. Dev Cell. 2007. PMID: 17543857 Review. - Constraints and limitations on the transcriptional response downstream of the Bicoid morphogen gradient.
Tran H, Walczak AM, Dostatni N. Tran H, et al. Curr Top Dev Biol. 2020;137:119-142. doi: 10.1016/bs.ctdb.2019.12.002. Epub 2020 Jan 17. Curr Top Dev Biol. 2020. PMID: 32143741 Review.
Cited by
- Image analysis and empirical modeling of gene and protein expression.
Trisnadi N, Altinok A, Stathopoulos A, Reeves GT. Trisnadi N, et al. Methods. 2013 Jul 15;62(1):68-78. doi: 10.1016/j.ymeth.2012.09.016. Epub 2012 Oct 24. Methods. 2013. PMID: 23104159 Free PMC article. - Evolutionary Design of Gene Networks: Forced Evolution by Genomic Parasites.
Spirov AV, Zagriychuk EA, Holloway DM. Spirov AV, et al. Parallel Process Lett. 2014 Jun;24(2):1440004. doi: 10.1142/S0129626414400040. Parallel Process Lett. 2014. PMID: 25558118 Free PMC article. - Accurate measurements of dynamics and reproducibility in small genetic networks.
Dubuis JO, Samanta R, Gregor T. Dubuis JO, et al. Mol Syst Biol. 2013;9:639. doi: 10.1038/msb.2012.72. Mol Syst Biol. 2013. PMID: 23340845 Free PMC article. - Temperature stress mediates decanalization and dominance of gene expression in Drosophila melanogaster.
Chen J, Nolte V, Schlötterer C. Chen J, et al. PLoS Genet. 2015 Feb 26;11(2):e1004883. doi: 10.1371/journal.pgen.1004883. eCollection 2015. PLoS Genet. 2015. PMID: 25719753 Free PMC article. - Diverse Spatial Expression Patterns Emerge from Unified Kinetics of Transcriptional Bursting.
Zoller B, Little SC, Gregor T. Zoller B, et al. Cell. 2018 Oct 18;175(3):835-847.e25. doi: 10.1016/j.cell.2018.09.056. Cell. 2018. PMID: 30340044 Free PMC article.
References
- Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. - PubMed
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. - PubMed
- Sollars V, Lu X, Xiao L, Wang X, Garfinkel MD, et al. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat Genet. 2002;33:70–74. - PubMed
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila . Nature. 1980;287:795–801. - PubMed
MeSH terms
Substances
Grants and funding
- R01 GM072022/GM/NIGMS NIH HHS/United States
- R01 RR007801/RR/NCRR NIH HHS/United States
- R01 RR007801-18/RR/NCRR NIH HHS/United States
- R56 GM072022/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous