A pre-S gene chip to detect pre-S deletions in hepatitis B virus large surface antigen as a predictive marker for hepatoma risk in chronic hepatitis B virus carriers - PubMed (original) (raw)
A pre-S gene chip to detect pre-S deletions in hepatitis B virus large surface antigen as a predictive marker for hepatoma risk in chronic hepatitis B virus carriers
Fan-Ching Shen et al. J Biomed Sci. 2009.
Abstract
Background: Chronic hepatitis B virus (HBV) infection is an important cause of hepatocellular carcinoma (HCC) worldwide. The pre-S1 and -S2 mutant large HBV surface antigen (LHBS), in which the pre-S1 and -S2 regions of the LHBS gene are partially deleted, are highly associated with HBV-related HCC.
Methods: The pre-S region of the LHBS gene in two hundred and one HBV-positive serum samples was PCR-amplified and sequenced. A pre-S oligonucleotide gene chip was developed to efficiently detect pre-S deletions in chronic HBV carriers. Twenty serum samples from chronic HBV carriers were analyzed using the chip.
Results: The pre-S deletion rates were relatively low (7%) in the sera of patients with acute HBV infection. They gradually increased in periods of persistent HBV infection: pre-S mutation rates were 37% in chronic HBV carriers, and as high as 60% in HCC patients. The Pre-S Gene Chip offers a highly sensitive and specific method for pre-S deletion detection and is less expensive and more efficient (turnaround time 3 days) than DNA sequencing analysis.
Conclusion: The pre-S1/2 mutants may emerge during the long-term persistence of the HBV genome in carriers and facilitate HCC development. Combined detection of pre-S mutations, other markers of HBV replication, and viral titers, offers a reliable predictive method for HCC risks in chronic HBV carriers.
Figures
Figure 1
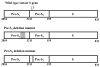
Representatives of the wild-type, pre-S1, and pre-S2 mutant LHBS gene. The shaded boxes are the regions deleted in the pre-S1 and pre-S2 mutant LHBS. The numbers on the bottom of the gene indicate the pre-S1, S2, and S regions of the LHBS gene in nucleotide order. Nucleotide 1 is the start of the circular genome and the numbers go clockwise. The last nt number is 3221. Here only the S gene, which spans the start of the genome, is shown. The arrow at the top of the diagram indicates the start (nt 1) of the HBV genome.
Figure 2
The Pre-S Gene Chip. The chip (7 mm (H) × 10 mm (W)) contains 42 oligonucleotide probes spanning the pre-S regions. The target region of each probe is indicated in the order of the nucleotides below the chip. The probes with extension numbers (e.g., 8-1, 8-2,... 8-6) are the redundant probes that target the same regions as the primary probes (e.g., 8) do.
Figure 3
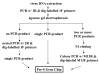
Working scheme of Pre-S Gene Chip analysis. The virus DNA is extracted from the patient's blood or liver tissue. The pre-S region is PCR-amplified using PCR-1R and 5'-dig-labeled 1F primers. The PCR products are visualized using agarose gel electrophoresis and ethidium bromide staining. In cases where no PCR products are seen---perhaps because of a low HBV DNA titer---nested PCR using PCR-2R and 5'-dig-labeled 1F primers is done. When only a single PCR product is seen, the DNA product is directly subjected to chip hybridization. However, when two or more different types of pre-S PCR products are seen in agarose gel, the products are first directed to TA cloning. Multiple plasmid clones are then analyzed using colony PCR with M13R and 5'-dig-labeled 13F primers. These PCR products are then analyzed using the Pre-S Gene Chip.
Figure 4
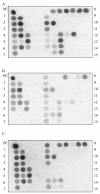
Results of the Pre-S Gene Chip analysis. A: The wild-type LHBS gene. B: The pre-S1 mutant LHBS, with nt 3044-3103 of the HBV genome deleted. The signals on probes 6 (nt 3045-3071) and 7 (nt 3074-3103) are negative. C: The pre-S2 mutant LHBS, with nt 3-57 of the HBV genome deleted. The signals on probes 12 (nt 3-32) and 13 (nt 33-62) are negative.
Similar articles
- Different pre-S deletion patterns and their association with hepatitis B virus genotypes.
Chen BF. Chen BF. World J Gastroenterol. 2016 Sep 21;22(35):8041-9. doi: 10.3748/wjg.v22.i35.8041. World J Gastroenterol. 2016. PMID: 27672298 Free PMC article. - Hepatitis B virus pre-S2 mutant large surface protein inhibits DNA double-strand break repair and leads to genome instability in hepatocarcinogenesis.
Hsieh YH, Chang YY, Su IJ, Yen CJ, Liu YR, Liu RJ, Hsieh WC, Tsai HW, Wang LH, Huang W. Hsieh YH, et al. J Pathol. 2015 Jul;236(3):337-47. doi: 10.1002/path.4531. Epub 2015 Apr 22. J Pathol. 2015. PMID: 25775999 - Association of hepatitis B virus pre-S deletions with the development of hepatocellular carcinoma in chronic hepatitis B.
Yeung P, Wong DK, Lai CL, Fung J, Seto WK, Yuen MF. Yeung P, et al. J Infect Dis. 2011 Mar 1;203(5):646-54. doi: 10.1093/infdis/jiq096. Epub 2011 Jan 12. J Infect Dis. 2011. PMID: 21227916 Free PMC article. - Hepatitis B virus pre-S/S variants in liver diseases.
Chen BF. Chen BF. World J Gastroenterol. 2018 Apr 14;24(14):1507-1520. doi: 10.3748/wjg.v24.i14.1507. World J Gastroenterol. 2018. PMID: 29662289 Free PMC article. Review. - Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis.
Wang HC, Huang W, Lai MD, Su IJ. Wang HC, et al. Cancer Sci. 2006 Aug;97(8):683-8. doi: 10.1111/j.1349-7006.2006.00235.x. Cancer Sci. 2006. PMID: 16863502 Free PMC article. Review.
Cited by
- Association of Low Serum Albumin Level with Higher Hepatocellular Carcinoma Recurrence in Patients with Hepatitis B Virus Pre-S2 Mutant after Curative Surgical Resection.
Jeng LB, Li TC, Hsu SC, Chan WL, Teng CF. Jeng LB, et al. J Clin Med. 2021 Sep 16;10(18):4187. doi: 10.3390/jcm10184187. J Clin Med. 2021. PMID: 34575311 Free PMC article. - Increased infiltration of regulatory T cells in hepatocellular carcinoma of patients with hepatitis B virus pre-S2 mutant.
Teng CF, Li TC, Wang T, Liao DC, Wen YH, Wu TH, Wang J, Wu HC, Shyu WC, Su IJ, Jeng LB. Teng CF, et al. Sci Rep. 2021 Jan 13;11(1):1136. doi: 10.1038/s41598-020-80935-5. Sci Rep. 2021. PMID: 33441885 Free PMC article. - Molecular characterization of hepatitis B virus among chronic hepatitis B patients from Pointe Noire, Republic of Congo.
Angounda BM, Ngouloubi GH, Dzia AB, Boumba LMA, Baha W, Moukassa D, Ahombo G, Ennaji MM, Ibara JR. Angounda BM, et al. Infect Agent Cancer. 2016 Sep 15;11:51. doi: 10.1186/s13027-016-0088-3. eCollection 2016. Infect Agent Cancer. 2016. PMID: 27651827 Free PMC article. - Molecular imprinting resonance light scattering nanoprobes based on pH-responsive metal-organic framework for determination of hepatitis A virus.
Luo L, Zhang F, Chen C, Cai C. Luo L, et al. Mikrochim Acta. 2020 Jan 18;187(2):140. doi: 10.1007/s00604-020-4122-1. Mikrochim Acta. 2020. PMID: 31955258 - The emerging role of hepatitis B virus pre-S2 deletion mutant proteins in HBV tumorigenesis.
Su IJ, Wang LH, Hsieh WC, Wu HC, Teng CF, Tsai HW, Huang W. Su IJ, et al. J Biomed Sci. 2014 Oct 15;21(1):98. doi: 10.1186/s12929-014-0098-7. J Biomed Sci. 2014. PMID: 25316153 Free PMC article. Review.
References
- Parkin DM, Whelan SL, Ferlay J, Storm H. IARC Cancer Base No. 7. WHO Press; 2005. Cancer Incidence in Five Continents.
- Bosch FX. In: Liver Cancer. Okuda K, Tabor E, editor. Churchill Livingstone, New York; 1997. Global epidemiology of hepatocellular carcinoma; pp. 13–28.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical