New insights into BAR domain-induced membrane remodeling - PubMed (original) (raw)
New insights into BAR domain-induced membrane remodeling
Gary S Ayton et al. Biophys J. 2009.
Abstract
Mesoscopic simulations and electron microscopy of N-BAR domain-induced liposome remodeling are used to characterize the process of liposome tubulation and vesiculation. The overall process of membrane remodeling is found to involve complex couplings among the N-BAR protein density, the degree of N-BAR oligomerization, and the membrane density. A comparison of complex remodeled liposome structures from mesoscopic simulations with those measured by electron microscopy experiments suggests that the process of membrane remodeling can be described via an appropriate mesoscopic free energy framework. Liposome remodeling more representative of F-BAR domains is also presented within the mesoscopic simulation framework.
Figures
Figure 1
(a) Snapshot of a putative N-BAR oligomerization structure on an atomistic membrane. (b) Square slab of EM2 membrane, 250 nm2 in area, and is prepared with a constant N-BAR density and membrane composition. Panels _b_-1–6 follow the membrane through the remodeling process. The arrows designate the local spontaneous curvature directions; the intersecting lines show the local curvature directions on the EM2 membrane.
Figure 2
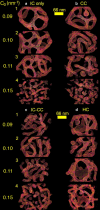
Tubulation and vesiculation of an EM2 liposome. The N-BAR spontaneous curvature magnitude is given in the far left column, starting from _C_0 = 0.09 nm−1, then _C_0 = 0.10 nm−1, _C_0 = 0.11 nm−1, and _C_0 = 0.15 nm−1. Panel a employs IC, panel b employs CC, panel c uses a combined 50:50 superposition of IC and CC, and panel d is for HC.
Figure 3
A comparison of structures obtained from simulation (left panel) and experimental electron micrographs (right panel). The boxes select out similar structural motifs found in both the simulations and experiment. Density variations in the effective outer leaflet of the EM2 membrane are shown in the smaller images (pink/amber online); where the pink regions have a lower outer bilayer leaflet lipid density. The N-BAR oligomerization fields as given by the EM2 nT vectors are shown in the larger images (blue-white online); darker regions have a relatively larger N-BAR density. In panels a_–_c, IC was used and the oligomerization strength, ΛO, was varied as in Eq. 11. Image c was obtained by removing the mesoscopic solvent after 200 ns of mesoscopic simulation and continuing the simulation in a solvent-free mode. The electron micrographs in panels a and b correspond to endophilin whereas panel c is amphiphysin N-BARs. Virtually indistinguishable tubule diameters were found for both systems.
Figure 4
Final F-BAR tubulation structure from the EM2 model based on Eqs. 1 and 11, with parameters chosen for F-BARs. An initial 500-nm-diameter liposome was used.
Similar articles
- Membrane remodeling from N-BAR domain interactions: insights from multi-scale simulation.
Ayton GS, Blood PD, Voth GA. Ayton GS, et al. Biophys J. 2007 May 15;92(10):3595-602. doi: 10.1529/biophysj.106.101709. Epub 2007 Feb 26. Biophys J. 2007. PMID: 17325001 Free PMC article. - Understanding the role of amphipathic helices in N-BAR domain driven membrane remodeling.
Cui H, Mim C, Vázquez FX, Lyman E, Unger VM, Voth GA. Cui H, et al. Biophys J. 2013 Jan 22;104(2):404-11. doi: 10.1016/j.bpj.2012.12.006. Biophys J. 2013. PMID: 23442862 Free PMC article. - Amphipathic motifs in BAR domains are essential for membrane curvature sensing.
Bhatia VK, Madsen KL, Bolinger PY, Kunding A, Hedegård P, Gether U, Stamou D. Bhatia VK, et al. EMBO J. 2009 Nov 4;28(21):3303-14. doi: 10.1038/emboj.2009.261. Epub 2009 Oct 8. EMBO J. 2009. PMID: 19816406 Free PMC article. - Membrane curvature: how BAR domains bend bilayers.
Zimmerberg J, McLaughlin S. Zimmerberg J, et al. Curr Biol. 2004 Mar 23;14(6):R250-2. doi: 10.1016/j.cub.2004.02.060. Curr Biol. 2004. PMID: 15043839 Review. - Higher-order assemblies of BAR domain proteins for shaping membranes.
Suetsugu S. Suetsugu S. Microscopy (Oxf). 2016 Jun;65(3):201-10. doi: 10.1093/jmicro/dfw002. Epub 2016 Feb 15. Microscopy (Oxf). 2016. PMID: 26884618 Review.
Cited by
- A bacterial membrane sculpting protein with BAR domain-like activity.
Phillips DA, Zacharoff LA, Hampton CM, Chong GW, Malanoski AP, Metskas LA, Xu S, Bird LJ, Eddie BJ, Miklos AE, Jensen GJ, Drummy LF, El-Naggar MY, Glaven SM. Phillips DA, et al. Elife. 2021 Oct 13;10:e60049. doi: 10.7554/eLife.60049. Elife. 2021. PMID: 34643180 Free PMC article. - Mechanism of membrane curvature sensing by amphipathic helix containing proteins.
Cui H, Lyman E, Voth GA. Cui H, et al. Biophys J. 2011 Mar 2;100(5):1271-9. doi: 10.1016/j.bpj.2011.01.036. Biophys J. 2011. PMID: 21354400 Free PMC article. - Application of a free-energy-landscape approach to study tension-dependent bilayer tubulation mediated by curvature-inducing proteins.
Tourdot RW, Ramakrishnan N, Baumgart T, Radhakrishnan R. Tourdot RW, et al. Phys Rev E Stat Nonlin Soft Matter Phys. 2015 Oct;92(4):042715. doi: 10.1103/PhysRevE.92.042715. Epub 2015 Oct 29. Phys Rev E Stat Nonlin Soft Matter Phys. 2015. PMID: 26565280 Free PMC article. - Membrane tubule formation by banana-shaped proteins with or without transient network structure.
Noguchi H. Noguchi H. Sci Rep. 2016 Feb 11;6:20935. doi: 10.1038/srep20935. Sci Rep. 2016. PMID: 26863901 Free PMC article. - Systems biology and physical biology of clathrin-mediated endocytosis.
Ramanan V, Agrawal NJ, Liu J, Engles S, Toy R, Radhakrishnan R. Ramanan V, et al. Integr Biol (Camb). 2011 Aug;3(8):803-15. doi: 10.1039/c1ib00036e. Epub 2011 Jul 26. Integr Biol (Camb). 2011. PMID: 21792431 Free PMC article. Review.
References
- Peter B.J., Kent H.M., Mills I.G., Vallis Y., Butler P.J.G. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. - PubMed
- Gallop J.L., McMahon H.T. BAR domains and membrane curvature: bringing your curves to the BAR. Biochem. Soc. Symp. 2005;72:223–231. - PubMed
- Zimmerberg J., McLaughlin S. Membrane curvature: how BAR domains bend bilayers. Curr. Biol. 2004;14:R250–R252. - PubMed
- Futterer K., Machesky L.M. “Wunder” F-BAR domains: going from pits to vesicles. Cell. 2007;129:655–657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM063796/GM/NIGMS NIH HHS/United States
- R21 DA024101/DA/NIDA NIH HHS/United States
- R01-GM063796/GM/NIGMS NIH HHS/United States
- R21-DA024101/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases