Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly - PubMed (original) (raw)
Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly
Paul Chang et al. Mol Biol Cell. 2009 Nov.
Abstract
Poly(ADP-ribose) (pADPr), made by PARP-5a/tankyrase-1, localizes to the poles of mitotic spindles and is required for bipolar spindle assembly, but its molecular function in the spindle is poorly understood. To investigate this, we localized pADPr at spindle poles by immuno-EM. We then developed a concentrated mitotic lysate system from HeLa cells to probe spindle pole assembly in vitro. Microtubule asters assembled in response to centrosomes and Ran-GTP in this system. Magnetic beads coated with pADPr, extended from PARP-5a, also triggered aster assembly, suggesting a functional role of the pADPr in spindle pole assembly. We found that PARP-5a is much more active in mitosis than interphase. We used mitotic PARP-5a, self-modified with pADPr chains, to capture mitosis-specific pADPr-binding proteins. Candidate binding proteins included the spindle pole protein NuMA previously shown to bind to PARP-5a directly. The rod domain of NuMA, expressed in bacteria, bound directly to pADPr. We propose that pADPr provides a dynamic cross-linking function at spindle poles by extending from covalent modification sites on PARP-5a and NuMA and binding noncovalently to NuMA and that this function helps promote assembly of exactly two poles.
Figures
Figure 1.
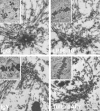
Ultrastructural localization of pADPr in the mitotic spindle. HeLa cells were permeabilized and fixed using a periodate/bis-amine/aldehdye protocol and then stained with two different antibodies to pADPr raised against distinct antigens (LP9610, BD Biosciences or anti-pADPr chicken, Tulip BioLabs) and control IgG or control IgY, followed by 5-nm gold conjugated to secondary antibody. Cells were then postfixed, embedded, and sectioned. A few gold particles were observed in sections from control IgG- or IgY-stained cells (left). Anti-pADPr staining (LP96–10 or pADPr) resulted in many gold particles near the spindle pole, particularly in the region between the centrosome and the spindle proper, where minus ends of spindle microtubule converge. Scale bar, 1 μm.
Figure 2.
Reconstitution of aspects of spindle pole assembly in concentrated mitotic lysate. To study pADPr contribution to spindle pole assembly, we developed a human cell extract system to recapitulate spindle pole assembly. In all cases examined, the human cell extracts behaved similar to Xenopus laevis egg extract spindle pole assembly reactions. (a) Centrosome induced asters. Concentrated lysates were prepared from mitotic HeLa cells expressing GFP-PARP-5a (top row) or GFP-NuMA (bottom row) and supplemented with Alexa 594 tubulin and purified mitotic centrosomes. Extracts were imaged after 30 min. Microtubules (MT) are shown in left panels and Merge, and PARP-5a-GFP or NuMA-GFP in the middle panels, and Merge in the right panels). Note assembly of asters that recruit the known spindle pole proteins PARP-5a and NuMA to their foci. In merge MTs are shown in green, and red represents the GFP-fusion. (b) Ran-GTP induced asters. The experiment was performed as in panel a, except that 1 mg/ml RanQ69L was added in place of centrosomes. Note the abundant asters. The Ran asters are less focused than centrosome-induced asters, as judged by PARP-5a and NuMA fluorescence. In merge MTs are shown in green, and red represents the GFP-fusion. (c) Examples of structures that assembled when mitotic chromosomes alone (left) or mitotic chromosomes and centrosomes (right) were incubated in lysate. Chromosomes alone induced some asters. When chromosomes and centrosomes were mixed, spindle-like assemblies containing dense microtubule bundles were observed. Microtubules are shown in green and DNA in blue.
Figure 3.
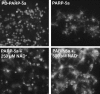
Aster assembly trigged by pADPr on beads. Magnetic beads coated with PARP-5a lacking catalytic activity (PD-PARP-5a) or PARP-5a decorated with pADPr (PARP-5a-pADPr) were added to concentrated mitotic lysate containing Alexa 594 tubulin and 1 μM ADP-ADHP to inhibit PARG activity. Note that pADPr-coated beads induced assembly of microtubule asters. Addition of extra NAD+ (+250 μM NAD+ and + 500 μM NAD+) to the lysate to promote additional pADPr polymerization increased the density of asters and the density of microtubules within them. Microtubule appeared not to emanate directly from the pADPr-coated beads; rather they assembled into asters that associated laterally with beads.
Figure 4.
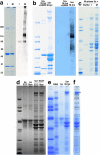
Composition of PARP-5a-pADPr beads and identification of mitotic pADPr-binding proteins. (a) PARP-5a activity is up-regulated during mitosis. GFP-PARP-5a was isolated from lysates of asynchronous (I) and mitosis-arrested (M) HeLa S3 cells using magnetic beads (see Materials and Methods). Beads were washed with high-salt buffer and analyzed by SDS-PAGE and Coomassie blue staining (left) or anti-pADPr Western blot (right). Note much greater pADPr labeling of the M sample. This includes a strong band at the position corresponding to the well of the gel, suggesting that heavily pADPr-labeled proteins failed to enter the gel. The beads shown in the M lane are typical of those used in Figure 3. The numbers to the left show the molecular weights (kDa) of the markers used in all the gels. (b) Identification of pADPr-binding proteins. Beads like those shown in panel a (M) were incubated with mitotic lysate at 4°C and washed with 0.3 M NaCl buffer. These beads were split in two; one-half was analyzed without further treatment (5a + M-lys), the other was treated with a pADPr glycohydrolase (ARH3) and washed (5a + M-lys + ARH3). Note a number of bands that are strongly labeled with Coomassie or anti-pADPr in the lane not treated with glycohydrolase. Most of these were removed by glycohydrolase, indicating they bind to pADPr, and not the beads themselves, or by unmodified PARP-5a. (c) Cell cycle specificity of pADPr-binding proteins. PARP-5a-pADPr beads were made in mitotic lysate and washed with 500 mM NaCl in lysate buffer and then incubated with buffer alone, lysate from asynchronous (I), or mitosis-arrested (M) cells. Note that some of the heaviest bands are similar between the two cell cycle states, but in general the profiles are very different. (d) Drug specificity and requirement for NAD+. pADPr-binding proteins were isolated from cells arrested in mitosis using drugs that inhibit kinesin-5, _S_-tritl-
l
-cysteine (STC), or microtubules, nocodazole (NOC). As another test of pADPr requirement, NAD+ was omitted from the both the initial and second extract incubations (left lanes) or added at 500 μM (right lanes). Note that omission of NAD+ caused loss of most of the candidate pADPr-binding proteins. The profile of these proteins was nearly identical for the two drug arrests. (e) Comparison of pADPr and RNA-binding proteins in mitotic lysate. GFP-PARP-5a-pADPr beads were prepared as in panel a. The gel lane labeled 5a shows these beads alone. RNA beads were prepared by incubating biotinylated RNA (Ambion) with streptavidin beads. Both beads were incubated with mitotic lysate, washed, and analyzed by SDS-PAGE. Note that pADPr-coated beads recruited a similar band pattern to b, c (M), and d. RNA beads recruited what appeared to be some similar proteins by band mobility, but that had an overall very different protein profile. (f) Example of band cutting for LC/MS analysis. The boxes indicate bands from the rightmost lane in the gel shown in panel e that were excised for LC/MS analysis. This type of analysis was performed twice, in addition to some LC/MS runs on total pADPr-binding proteins. See Table 1 for the identity of the most abundant protein in each excised band.
Figure 5.
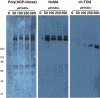
Recruitment of NuMA and CH-TOG to pADPr beads at different polymer levels. Molecular weights shown at left. GFP-PARP-5a-pADPr beads were prepared as in panel a and added to aliquots of mitotic lysate for recruitment of binding proteins. The lysates were supplemented with the indicated concentration of NAD+ and the PARG inhibitor ADP-ADHP (1 μM) except for the zero NAD+, where it was omitted. Beads were washed and blotted for pADPr, NuMA, and CH-TOG. Increasing NAD+ concentrations resulted in increased levels of pADPr on the beads, as revealed by the pADPr immunoblot. NuMA was recruited equally to the beads at all nonzero concentrations of NAD+, whereas CH-TOG recruitment increased with increasing concentrations of NAD+ and increasing amounts of pADPr on the beads.
Figure 6.
NuMA rod domain binds directly to p(ADPr). Molecular weights are shown at left. NuMA was expressed in bacteria as three nonoverlapping fragments tagged with His6. Top, these corresponded approximately to the main structural (terminal) domains in NuMA: N-term., aa 1–216; C-term., aa1701–2102; and Rod domain, aa. 217-1701. Bottom, fragments were incubated with PARP-5a-pADPr (5a) bound to magnetic beads and then were washed and analyzed by SDS-PAGE. In all cases I refers to input and p to the washed PARP-5a-input samples. Only the Rod domain bound to the beads. For lane 8, beads from a Rod domain incubation were treated with the glycohydrolase ARH3 and washed. Note Rod domain binding to PARP-5a-pARPr was lost, indicating it binds to the pADPr on the beads and not to the beads themselves or to PARP-5a polypeptide.
Figure 7.
Model for pADPr function at spindle poles. Microtubules are shown as green lines with their polarity indicated by arrowheads. NuMA (yellow pentagon) and minus-directed motors (not shown) are sufficient to locally cluster minus ends. PARP-5a (blue rectangle) is recruited to minus-end clusters by protein–protein interaction with NuMA. Polymerization of pADPr (red lines) from priming sites on PARP-5a and NuMA recruits more NuMA, which promotes further condensation of minus-end clusters into two poles. Note that this diagram is drawn to illustrate a conceptual hierarchy of interactions, not a temporal sequence of events. The blow-up illustrates that the pADPr chains attach covalently to PARP-5a and NuMA (red dots at end of polymer) and interact noncovalently with NuMA.
Similar articles
- NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis.
Chang W, Dynek JN, Smith S. Chang W, et al. Biochem J. 2005 Oct 15;391(Pt 2):177-84. doi: 10.1042/BJ20050885. Biochem J. 2005. PMID: 16076287 Free PMC article. - Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function.
Chang P, Coughlin M, Mitchison TJ. Chang P, et al. Nat Cell Biol. 2005 Nov;7(11):1133-9. doi: 10.1038/ncb1322. Nat Cell Biol. 2005. PMID: 16244666 - Interaction of NuMA protein with the kinesin Eg5: its possible role in bipolar spindle assembly and chromosome alignment.
Iwakiri Y, Kamakura S, Hayase J, Sumimoto H. Iwakiri Y, et al. Biochem J. 2013 Apr 15;451(2):195-204. doi: 10.1042/BJ20121447. Biochem J. 2013. PMID: 23368718 - NuMA after 30 years: the matrix revisited.
Radulescu AE, Cleveland DW. Radulescu AE, et al. Trends Cell Biol. 2010 Apr;20(4):214-22. doi: 10.1016/j.tcb.2010.01.003. Trends Cell Biol. 2010. PMID: 20137953 Free PMC article. Review. - Role of NuMA in vertebrate cells: review of an intriguing multifunctional protein.
Sun QY, Schatten H. Sun QY, et al. Front Biosci. 2006 Jan 1;11:1137-46. doi: 10.2741/1868. Front Biosci. 2006. PMID: 16146802 Review.
Cited by
- A FRET-based high-throughput screening platform for the discovery of chemical probes targeting the scaffolding functions of human tankyrases.
Sowa ST, Vela-Rodríguez C, Galera-Prat A, Cázares-Olivera M, Prunskaite-Hyyryläinen R, Ignatev A, Lehtiö L. Sowa ST, et al. Sci Rep. 2020 Jul 23;10(1):12357. doi: 10.1038/s41598-020-69229-y. Sci Rep. 2020. PMID: 32704068 Free PMC article. - Tankyrase inhibition promotes a stable human naïve pluripotent state with improved functionality.
Zimmerlin L, Park TS, Huo JS, Verma K, Pather SR, Talbot CC Jr, Agarwal J, Steppan D, Zhang YW, Considine M, Guo H, Zhong X, Gutierrez C, Cope L, Canto-Soler MV, Friedman AD, Baylin SB, Zambidis ET. Zimmerlin L, et al. Development. 2016 Dec 1;143(23):4368-4380. doi: 10.1242/dev.138982. Epub 2016 Sep 22. Development. 2016. PMID: 27660325 Free PMC article. - A sePARate phase? Poly(ADP-ribose) versus RNA in the organization of biomolecular condensates.
Alemasova EE, Lavrik OI. Alemasova EE, et al. Nucleic Acids Res. 2022 Oct 28;50(19):10817-10838. doi: 10.1093/nar/gkac866. Nucleic Acids Res. 2022. PMID: 36243979 Free PMC article. Review. - Tankyrase-1 function at telomeres and during mitosis is regulated by Polo-like kinase-1-mediated phosphorylation.
Ha GH, Kim HS, Go H, Lee H, Seimiya H, Chung DH, Lee CW. Ha GH, et al. Cell Death Differ. 2012 Feb;19(2):321-32. doi: 10.1038/cdd.2011.101. Epub 2011 Aug 5. Cell Death Differ. 2012. PMID: 21818122 Free PMC article.
References
- Ahel I., Ahel D., Matsusaka T., Clark A. J., Pines J., Boulton S. J., West S. C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. - PubMed
- Becker B. E., Romney S. J., Gard D. L. XMAP215, XKCM1, NuMA, and cytoplasmic dynein are required for the assembly and organization of the transient microtubule array during the maturation of Xenopus oocytes. Dev. Biol. 2003;261:488–505. - PubMed
- Blower M. D., Nachury M., Heald R., Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. - PubMed
- Bradbury S., Stoward P. J. The specific cytochemical demonstration in the electron microscope of periodate-reactive mucosubstances and polysaccharides containing vic-glycol groups. Histochemie. 1967;11:71–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM070090/GM/NIGMS NIH HHS/United States
- R01 GM039565/GM/NIGMS NIH HHS/United States
- 5F32GM070090-02/GM/NIGMS NIH HHS/United States
- R37 GM039565/GM/NIGMS NIH HHS/United States
- GM39565/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous