Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc - PubMed (original) (raw)
Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc
H Wayne Peng et al. Genes Dev. 2009.
Abstract
The accurate control of cell proliferation and survival is critical for animal development. The Hippo tumor suppressor pathway regulates both of these parameters by controlling the nuclear availability of the transcriptional coactivator Yorkie (Yki), which regulates downstream target genes together with Scalloped (Sd), a DNA-binding protein. Here we provide evidence that Yki can also regulate target genes in conjunction with Homothorax (Hth) and Teashirt (Tsh), two DNA-binding transcription factors expressed in the uncommitted progenitor cells of the Drosophila eye imaginal disc. Clonal analyses demonstrate that Hth and Tsh promote cell proliferation and protect eye progenitor cells from apoptosis. Genetic epistasis experiments suggest that Hth and Tsh execute these functions with Yki, in part by up-regulating the microRNA bantam. A physical interaction between Hth and Yki can be detected in cell culture, and we show that Hth and Yki are bound to a DNA sequence approximately 14 kb upstream of the bantam hairpin in eye imaginal disc cells, arguing that this regulation is direct. These data suggest that the Hippo pathway uses different DNA-binding transcription factors depending on the cellular context. In the eye disc, Hth and Tsh provide spatial information to this pathway, promoting cell proliferation and survival in the progenitor domain.
Figures
Figure 1.
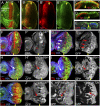
hth is required for cell survival in the eye progenitor domain. (A) A third instar eye disc stained for CycB (green) and Dac (red, which is expressed anterior to the MF) to highlight the progenitor domain and the initiation of differentiation, respectively. The position and direction of the MF is indicated. (B) A third instar eye disc stained for Hth (green) and Tsh (red). (C–E) Confocal cross-section of third instar eye discs stained for Dlg (green, all panels), Hth (red, C), Tsh (red, D), pH3 (red, E), and Elav (blue, E). The position of MF is indicated (yellow arrowheads). Hth is present in the anterior of the main epithelium, throughout the peripodial epithelium, and in posterior margin cells (arrow). Tsh is expressed only in the main epithelium, and extends closer to the MF compared with Hth. Dlg is used here as a cell membrane marker. (F,G) Survival of hthP2 (F) or neutral clones (G) (marked by the absence of GFP) in eye discs stained for Hth (blue) and Dlg (red, which marks the MF, arrowheads). hthP2 clones are not recovered anterior to the MF, but twin spots (bright patches in _F_′,_G_′) are recovered. (H–J) A few hthP2 clones (marked by the absence of GFP, green/white) are recovered anterior to the MF in a H99/+ background (H) or when the apoptosis inhibitor p35 is expressed (J), but not as readily as neutral control clones (I). (K,L) Comparison of the growth of hthP2 Minute+ (K) and hth+ Minute+ (L) clones made in a Minute+/_Minute_− background.
Figure 2.
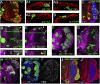
Hth and Tsh function together to induce proliferation. All panels show eye discs with positively marked (GFP+) clones. (A,B) Eye discs containing clones that ectopically express Tsh (A) or GFP (B), stained for GFP (green, to mark the clones) and Dlg (red). Compared with neutral clones (B), Tsh+ clones result in overgrowths (arrowheads) when they arise in the posterior margin (A1) or in the peripodial epithelium (A2), but not when they arise in the main epithelium (A3). (C) Eye disc containing clones that ectopically express Hth (marked with GFP, green), stained for Tsh (red) and Elav (blue). These clones do not result in overgrowths. (D,E) MARCM clones that ectopically express Tsh (D) or that ectopically express Tsh and are also mutant for hth (E), stained for Hth (purple) and GFP (to mark the clones). Tsh+ clones result in overgrowths (arrowheads in D), but Tsh+; hthP2 clones do not (arrowheads in E). _D_′ and _E_′ show confocal cross-sections indicated by the lines in D and E. (F,G) Eye discs containing Hth + Tsh clones (F) or neutral clones (G) stained for Hth (purple) and GFP (to mark the clones). Hth + Tsh clones always overgrow compared with control clones made in parallel. (H–J) Eye discs containing Hth + Tsh clones up-regulate CycB (purple, H) and pH3 (red/ white in I,_I_′), and repress Elav (blue, J,_J_′).
Figure 3.
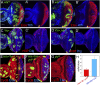
Codependency of the Hippo pathway and Hth + Tsh to induce proliferation in the eye progenitor domain. A_–_G all show eye discs with positively marked (GFP+) MARCM clones. (A,B) Eye discs with wtsP2 (A) and wtsP2 hthP2 (B) clones, stained for GFP (green, to mark the clones), Hth (blue), and Elav (red). wtsP2 clones grow well throughout the eye disc, whereas wtsP2 hthP2 clones are not recovered in the progenitor domain. (C,D) Eye discs with Yki+ (C) or Yki+; hthP2 (D) clones stained for GFP (green, to mark the clones), pH3 (red), or Dlg (blue). Yki+ clones grow well throughout the eye disc, whereas Yki+; hthP2 clones are not recovered in the progenitor domain. (E–G) Eye discs with Hth + Tsh (E), Hth + Tsh; ykiB5 (F), or ykiB5 (G) clones stained for GFP (green, to mark the clones), CycB (red), and Elav (blue). In E and F, the insets show only the Elav and CycB stains of the boxed regions. Note that Hth + Tsh clones, but not Hth + Tsh; ykiB5 clones, repress the differentiation marker Elav and up-regulate CycB. (H) The number of cells present in Hth + Tsh; ykiB5 clones (red bar; n = 55) and Hth + Tsh clones (blue bar; n = 58).
Figure 4.
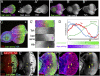
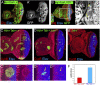
Control of bantam expression by Hth + Tsh. (A,B) Early to mid-third instar eye discs stained for the bantam sensor (green) and Hth (purple, A) or Tsh (purple, B). (C) Late third instar eye disc stained for Tsh (red), ban sensor (green), and Hth (blue). _C_′ shows a blow-up and individual channels for the region boxed in C. (D) Quantification of the Hth, Tsh, and ban sens stains in the regions boxed in C. bantam activity (purple gradient) is inferred to be the inverse for what is observed for the ban sensor (green gradient). (E) Eye disc containing clones expressing Hth + Tsh (absence of red, CD2) stained for the ban sensor (green/white in C,_C_′) and Elav (blue in C). Hth + Tsh clones repress both the very high levels of the ban sensor in the Elav+ cells and the intermediate ban sensor levels that are present close to the MF (arrowheads). (F) Eye disc containing hthP2 clones made in a H99/+ background stained for β-gal (red, absence of arm-lacZ expression marks the clones), Hth (blue), and the ban sensor (green). An increase in ban sensor levels compared with surrounding cells can be seen in three of three hthP2 clones that survived in the progenitor domain (arrows).
Figure 5.
Mutual dependence of bantam and Hth + Tsh for cell proliferation and survival. (A,B) Eye discs expressing bantam via eyeless-Gal4 (ey > ban) containing neutral clones (A) or hthP2 clones (B) stained for GFP (a clone marker), Elav (blue), and Dlg (red). A few hthP2 clones surviving in the progenitor domain are observed in B (arrows). (C–E) Eye discs containing Hth + Tsh clones (C), Hth + Tsh; banΔ1 clones (D), or banΔ1 clones (E) stained for CycB (red) and Elav (blue). _C_′, _C_″, _D_′, and _D_″ show enlargements of the boxed regions in C and D, respectively. Hth + Tsh; banΔ1 clones (D) grow more poorly compared with Hth + Tsh clones (C) made in parallel. Note also that Hth + Tsh; banΔ1 clones fail to repress Elav and do not activate CycB. (F) The number of cells present in Hth + Tsh; banΔ1 clones (red bar, n = 44) and Hth + Tsh clones (blue bar, n = 69).
Figure 6.
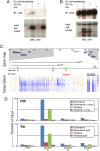
Molecular interactions between Hth, Yki, and the bantam locus. (A,B) Western blots of extracts from S2 cells that were transfected with the indicated expression plasmids. Blots were probed with anti-Hth (A) or anti-HA (B). The top panels show blots of immunoprecipitations with anti-HA (A) or anti-Hth (B), and the bottom panels show blots of total cell lysates. In A, Hth is coimmunoprecipitated with HA-Yki. In B, HA-Yki is coimmunoprecipitated with Hth. In A, the red arrows point to Hth detected in total cell lysates and anti-HA immunoprecipitate. In B, the red arrows point to HA-Yki detected in total cell lysates and anti-Hth immunoprecipitate. The asterisk in B indicates cross-reaction from the IgG heavy chain in the anti-Hth antiserum. (C) The top section shows a gene map in the vicinity of bantam; the regions deleted in two ban deletion mutants (banΔ1 and ban20) are indicated (Hipfner et al. 2002; Yang et al. 2009). The middle section reproduces data from the modENCODE project measuring the amount of transcription around the bantam locus; each row represents a different developmental time point (from embryogenesis to second instar) and provides a measure of amount of mRNA encoded by that genomic position; blue indicates high expression, and orange indicates low expression (see
for details). Regions A, B, and C indicate the positions of three of 11 primers used for ChIP experiments; based on the modENCODE transcription map, region A is close to an apparent start of bantam transcription and was the only amplicon that gave a positive ChIP signal. (D) The results of ChIP experiments using anti-Hth (top) and anti-Yki (bottom) antibodies to precipitate chromatin prepared from the indicated tissues. In the anti-Hth experiment, “comp” refers to the addition of a competitor peptide that specifically blocks binding to Hth (see the Materials and Methods for details). In the anti-Yki experiments, the control was unprogrammed IgG. Immunoprecipitated chromatin was assayed for the presence of regions A, B, or C or for a negative control locus, PDH. Data are presented as a percentage of the signal obtained relative to input chromatin. For both anti-Hth and anti-Yki, a significant signal was obtained only for region A.
Figure 7.
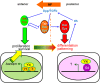
Model for the switch from proliferation to differentiation in the eye disc. Hth and Tsh are expressed in the progenitor domain where they repress retinal differentiation genes such as dac and eya and also up-regulate bantam expression in conjunction with Yki. Signals such as Dpp from the MF repress hth and tsh, allowing dac and eya to execute the differentiation program. In this region of the eye disc, Yki may work with other transcription factors such as Sd to regulate a different subset of Hippo pathway target genes.
Similar articles
- Yki/YAP, Sd/TEAD and Hth/MEIS control tissue specification in the Drosophila eye disc epithelium.
Zhang T, Zhou Q, Pignoni F. Zhang T, et al. PLoS One. 2011;6(7):e22278. doi: 10.1371/journal.pone.0022278. Epub 2011 Jul 19. PLoS One. 2011. PMID: 21811580 Free PMC article. - Divergent transcriptional regulatory logic at the intersection of tissue growth and developmental patterning.
Slattery M, Voutev R, Ma L, Nègre N, White KP, Mann RS. Slattery M, et al. PLoS Genet. 2013;9(9):e1003753. doi: 10.1371/journal.pgen.1003753. Epub 2013 Sep 5. PLoS Genet. 2013. PMID: 24039600 Free PMC article. - The retinal determination gene Dachshund restricts cell proliferation by limiting the activity of the Homothorax-Yorkie complex.
Brás-Pereira C, Casares F, Janody F. Brás-Pereira C, et al. Development. 2015 Apr 15;142(8):1470-9. doi: 10.1242/dev.113340. Epub 2015 Mar 19. Development. 2015. PMID: 25790852 - A role for Hipk in the Hippo pathway.
Heidary Arash E, Attisano L. Heidary Arash E, et al. Sci Signal. 2013 May 14;6(275):pe18. doi: 10.1126/scisignal.2004259. Sci Signal. 2013. PMID: 23674821 Review. - Signaling in the third dimension: the peripodial epithelium in eye disc development.
Atkins M, Mardon G. Atkins M, et al. Dev Dyn. 2009 Sep;238(9):2139-48. doi: 10.1002/dvdy.22034. Dev Dyn. 2009. PMID: 19623613 Free PMC article. Review.
Cited by
- Tshz1 Regulates Pancreatic β-Cell Maturation.
Raum JC, Soleimanpour SA, Groff DN, Coré N, Fasano L, Garratt AN, Dai C, Powers AC, Stoffers DA. Raum JC, et al. Diabetes. 2015 Aug;64(8):2905-14. doi: 10.2337/db14-1443. Epub 2015 Apr 27. Diabetes. 2015. PMID: 25918232 Free PMC article. - The BTB-zinc finger transcription factor abrupt acts as an epithelial oncogene in Drosophila melanogaster through maintaining a progenitor-like cell state.
Turkel N, Sahota VK, Bolden JE, Goulding KR, Doggett K, Willoughby LF, Blanco E, Martin-Blanco E, Corominas M, Ellul J, Aigaki T, Richardson HE, Brumby AM. Turkel N, et al. PLoS Genet. 2013;9(7):e1003627. doi: 10.1371/journal.pgen.1003627. Epub 2013 Jul 18. PLoS Genet. 2013. PMID: 23874226 Free PMC article. - Oxidized low-density lipoprotein promotes vascular endothelial cell dysfunction by stimulating miR-496 expression and inhibiting the Hippo pathway effector YAP.
Hu J, Liu T, Zhang Z, Xu Y, Zhu F. Hu J, et al. Cell Biol Int. 2019 May;43(5):528-538. doi: 10.1002/cbin.11120. Epub 2019 Mar 19. Cell Biol Int. 2019. PMID: 30811087 Free PMC article. - Master regulators in development: Views from the Drosophila retinal determination and mammalian pluripotency gene networks.
Davis TL, Rebay I. Davis TL, et al. Dev Biol. 2017 Jan 15;421(2):93-107. doi: 10.1016/j.ydbio.2016.12.005. Epub 2016 Dec 13. Dev Biol. 2017. PMID: 27979656 Free PMC article. Review. - Rapid diversification of homothorax expression patterns after gene duplication in spiders.
Turetzek N, Khadjeh S, Schomburg C, Prpic NM. Turetzek N, et al. BMC Evol Biol. 2017 Jul 14;17(1):168. doi: 10.1186/s12862-017-1013-0. BMC Evol Biol. 2017. PMID: 28709396 Free PMC article.
References
- Azpiazu N, Morata G. Function and regulation of homothorax in the wing imaginal disc of Drosophila. Development. 2000;127:2685–2693. - PubMed
- Bessa J, Casares F. Restricted teashirt expression confers eye-specific responsiveness to Dpp and Wg signals during eye specification in Drosophila. Development. 2005;132:5011–5020. - PubMed
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. - PubMed
- Campbell S, Inamdar M, Rodrigues V, Raghavan V, Palazzolo M, Chovnick A. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes & Dev. 1992;6:367–379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases