A cell surface interaction network of neural leucine-rich repeat receptors - PubMed (original) (raw)
A cell surface interaction network of neural leucine-rich repeat receptors
Christian Söllner et al. Genome Biol. 2009.
Abstract
Background: The vast number of precise intercellular connections within vertebrate nervous systems is only partly explained by the comparatively few known extracellular guidance cues. Large families of neural orphan receptor proteins have been identified and are likely to contribute to these recognition processes but due to the technical difficulty in identifying novel extracellular interactions of membrane-embedded proteins, their ligands remain unknown.
Results: To identify novel neural recognition signals, we performed a large systematic protein interaction screen using an assay capable of detecting low affinity extracellular protein interactions between the ectodomains of 150 zebrafish receptor proteins containing leucine-rich-repeat and/or immunoglobulin superfamily domains. We screened 7,592 interactions to construct a network of 34 cell surface receptor-ligand pairs that included orphan receptor subfamilies such as the Lrrtms, Lrrns and Elfns but also novel ligands for known receptors such as Robos and Unc5b. A quantitative biochemical analysis of a subnetwork involving the Unc5b and three Flrt receptors revealed a surprising quantitative variation in receptor binding strengths. Paired spatiotemporal gene expression patterns revealed dynamic neural receptor recognition maps within the developing nervous system, providing biological support for the network and revealing likely functions.
Conclusions: This integrated interaction and expression network provides a rich source of novel neural recognition pathways and highlights the importance of quantitative systematic extracellular protein interaction screens to mechanistically explain neural wiring patterns.
Figures
Figure 1
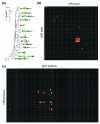
The leucine-rich repeat receptor family and its interactions in zebrafish. (a) Zebrafish LRR proteins were phylogenetically clustered into subfamilies using MegAlign (DNASTAR, Madison, WI, USA), and are shown as a phylogenetic tree, together with a schematic representation of their protein architecture. All the proteins shown were included in the protein-protein interaction screen. Protein domain abbreviations: LRR = leucine-rich repeat; LRRNT = leucine-rich repeat amino-terminal domain; LRRCT = leucine-rich repeat carboxy-terminal domain; IG = immunoglobulin superfamily domain; FN = fibronectin type III domain; TIR = Toll/interleukin-1 receptor homology domain; TM = transmembrane region; GPI = glycophosphatidylinositol anchor. (b) A binding grid showing all tested reciprocal interactions between the extracellular LRR proteins using AVEXIS. The baits are vertically ordered in correspondence to the tree shown in (a) and numbered as described in Additional data file 4. The preys are similarly ordered horizontally such that homophilic interactions are on the diagonal from top left to bottom right. Interactions identified by a red square were positive in both screens; blue squares were detected only once, but reciprocated. Baits 7, 8, 26 and 50 and prey 43 were expressed below the threshold required for the assay and were therefore not included in the screen. (c) A binding grid showing the interaction screen between the zebrafish LRR and IgSF receptor families. The 97 IgSF proteins are ordered horizontally according to their phylogenetic relationships and numbered as described in Additional data file 5; the 52 LRR proteins are similarly arranged vertically. Red and yellow squares indicate high and lower confidence interactions, respectively, as detailed in Additional data file 6.
Figure 2
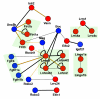
The extracellular LRR and IgSF neuroreceptor interaction network. Receptors belonging to the same paralogous subfamily are grouped and shaded within the network, and interactions classified according to confidence: thick lines = interaction detected in the primary screen and independent of bait/prey orientation; thin line = other detected interactions, including those that are orientation dependent - see Materials and methods and Additional data file 6 for full details. An orange line indicates that the interactions were validated using an independent technique, either surface plasmon resonance (Figure 3) or a bead-binding assay (Additional data file 2). IgSF-receptors = blue nodes, LRR-receptors = red nodes.
Figure 3
Interaction strengths between Unc5b and Flrt paralogs are surprisingly heterogeneous. (a) Equilibrium binding analysis of Unc5b and three Flrt paralogs. Different concentrations of purified, monomeric Unc5b-Cd4d3+4-6H were injected over streptavidin-coated flow cells upon which biotinylated baits - Flrt1a (1018 RU), Flrt1b (984 RU), Flrt3 (1027 RU) - and control Cd4d3+4 were immobilized. The amount of bound Unc5b was calculated by subtracting responses in the control flow cells from those in the Flrt-immobilized cells once equilibrium had been reached. Equilibrium dissociation constants (KDs) were obtained by fitting a non-linear binding curve to the data. To facilitate comparison, the binding responses were normalized by using the predicted Rmax from the fit to the data. (b) Kinetic analysis of the Unc5b-Flrt interactions. Off-rate constants (koff) were calculated by globally fitting a first order decay curve to the dissociation phase of three concentrations of Unc5b; half-lives (t1/2) were calculated as t1/2 = ln2/koff. Shown are the normalized, averaged values (error bars = ± 1 standard deviation, n = 3). On-rate constants (kon) were calculated in the same way using an association model and were > 1 × 105 M-1s-1 in all cases.
Figure 4
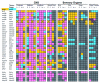
Genes encoding interacting receptors show compatible spatiotemporal expression. Genes encoding interacting receptors are paired (gene 1, gene 2) and listed vertically; homophilic interactions were treated separately below. The expression in anatomically distinct regions of the nervous system at different stages of embryonic development is indicated by appropriate shading within the grid. Expression key: gene 1 only = gold; gene 2 only = blue; co-expression = pink; no expression = grey; hatched = both expressed at the same stage outside the nervous system but not in identical or neighboring tissues; cross = expression not determined for one of the two genes. Stages: S = 14 to 19 somites; P = Prim5; Lp = Long-pec; L = Larval (4 to 5 days post-fertilization). CNS = central nervous system.
Figure 5
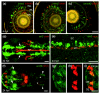
Two-color wholemount in situ hybridization of interacting neuroreceptors. (a-c) Single optical sections showing largely non-overlapping expression of the Lrrtm genes within the inner nuclear and ganglion cell layers of 4 days post-fertilization zebrafish retinae. Note that the confluent yellow staining within the lens represents background auto-fluorescence in both channels. (d-e) Neuron-glia interactions. (d) Dorsal view of the head region of a 32 hour post-fertilization (hpf) zebrafish embryo: vasn (red) is expressed in the most ventral part of the spinal cord in the medial floor plate cells (FP). islr2 (green) is expressed in fore-, mid- and hindbrain neurons; note that the midbrain neurons are in direct contact with the floor plate (arrows). (e) Lateral view of the developing spinal cord of a 24 hpf zebrafish embryo showing discrete cells within the spinal cord (SC) that are directly adjacent but dorsal to the floor plate. (f-i) Dorsal views of a 24 hpf zebrafish embryo showing expression of unc5b (green) and its interacting partner flrt1b (red). (f) unc5b is expressed in the dorsal retina (arrows) and the ear (arrowheads), flrt1b in the dorsal regions of the lateral midbrain and mid-hindbrain boundary; expression is also detectable in the vestibulo-acoustic ganglion (asterisks). (g-i) Higher magnification of the forebrain showing that unc5b is also expressed in the medial part of the olfactory bulb (g) where it overlaps with the flrt1b staining (h) in the olfactory bulb (OB) and olfactory epithelium (OE) (i). Scale bars: 50 μm (a-c); 80 μm (d); 40 μm (e); and 50 μm (f-i). GCL = retinal ganglion cell layer; INL = inner nuclear layer; IPL = inner plexiform layer; L = lens; OB = olfactory bulb; OE = olfactory epithelium.
Similar articles
- Construction of a large extracellular protein interaction network and its resolution by spatiotemporal expression profiling.
Martin S, Söllner C, Charoensawan V, Adryan B, Thisse B, Thisse C, Teichmann S, Wright GJ. Martin S, et al. Mol Cell Proteomics. 2010 Dec;9(12):2654-65. doi: 10.1074/mcp.M110.004119. Epub 2010 Aug 27. Mol Cell Proteomics. 2010. PMID: 20802085 Free PMC article. - G-protein-coupled estrogen receptor 1 is involved in brain development during zebrafish (Danio rerio) embryogenesis.
Shi Y, Liu X, Zhu P, Li J, Sham KW, Cheng SH, Li S, Zhang Y, Cheng CH, Lin H. Shi Y, et al. Biochem Biophys Res Commun. 2013 May 24;435(1):21-7. doi: 10.1016/j.bbrc.2013.03.130. Epub 2013 Apr 11. Biochem Biophys Res Commun. 2013. PMID: 23583372 - Essential role for the d-Asb11 cul5 Box domain for proper notch signaling and neural cell fate decisions in vivo.
Sartori da Silva MA, Tee JM, Paridaen J, Brouwers A, Runtuwene V, Zivkovic D, Diks SH, Guardavaccaro D, Peppelenbosch MP. Sartori da Silva MA, et al. PLoS One. 2010 Nov 19;5(11):e14023. doi: 10.1371/journal.pone.0014023. PLoS One. 2010. PMID: 21124961 Free PMC article. - Identification and characterization of zebrafish semaphorin 6D.
Kimura M, Taniguchi M, Mikami Y, Masuda T, Yoshida T, Mishina M, Shimizu T. Kimura M, et al. Biochem Biophys Res Commun. 2007 Nov 23;363(3):762-8. doi: 10.1016/j.bbrc.2007.09.038. Epub 2007 Sep 20. Biochem Biophys Res Commun. 2007. PMID: 17897628 - Neurexins, neuroligins and LRRTMs: synaptic adhesion getting fishy.
Wright GJ, Washbourne P. Wright GJ, et al. J Neurochem. 2011 Jun;117(5):765-78. doi: 10.1111/j.1471-4159.2010.07141.x. Epub 2011 Jan 19. J Neurochem. 2011. PMID: 21155806 Free PMC article. Review.
Cited by
- No evidence for a direct extracellular interaction between human Fc receptor-like 3 (MAIA) and the sperm ligand IZUMO1.
Bianchi E, Jiménez-Movilla M, Cots-Rodríguez P, Viola C, Wright GJ. Bianchi E, et al. Sci Adv. 2024 Feb 23;10(8):eadk6352. doi: 10.1126/sciadv.adk6352. Epub 2024 Feb 21. Sci Adv. 2024. PMID: 38381819 Free PMC article. - Clinical significance of UNC5B expression in bladder cancer.
Liu J, Zhang Z, Li ZH, Kong CZ. Liu J, et al. Tumour Biol. 2013 Aug;34(4):2099-108. doi: 10.1007/s13277-012-0532-y. Epub 2012 Oct 2. Tumour Biol. 2013. PMID: 23055195 - Proteolytically released Lasso/teneurin-2 induces axonal attraction by interacting with latrophilin-1 on axonal growth cones.
Vysokov NV, Silva JP, Lelianova VG, Suckling J, Cassidy J, Blackburn JK, Yankova N, Djamgoz MB, Kozlov SV, Tonevitsky AG, Ushkaryov YA. Vysokov NV, et al. Elife. 2018 Nov 20;7:e37935. doi: 10.7554/eLife.37935. Elife. 2018. PMID: 30457553 Free PMC article. - Development of an antigen microarray for high throughput monoclonal antibody selection.
Staudt N, Müller-Sienerth N, Wright GJ. Staudt N, et al. Biochem Biophys Res Commun. 2014 Mar 21;445(4):785-90. doi: 10.1016/j.bbrc.2013.12.033. Epub 2014 Jan 25. Biochem Biophys Res Commun. 2014. PMID: 24472540 Free PMC article. - Targeted cyclooxygenase-2 inhibiting nanomedicine results in pain-relief and differential expression of the RNA transcriptome in the dorsal root ganglia of injured male rats.
Stevens AM, Saleem M, Deal B, Janjic J, Pollock JA. Stevens AM, et al. Mol Pain. 2020 Jan-Dec;16:1744806920943309. doi: 10.1177/1744806920943309. Mol Pain. 2020. PMID: 32762277 Free PMC article.
References
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials