Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India - PubMed (original) (raw)
Case Reports
. 2009 Dec;53(12):5046-54.
doi: 10.1128/AAC.00774-09. Epub 2009 Sep 21.
Affiliations
- PMID: 19770275
- PMCID: PMC2786356
- DOI: 10.1128/AAC.00774-09
Case Reports
Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India
Dongeun Yong et al. Antimicrob Agents Chemother. 2009 Dec.
Abstract
A Swedish patient of Indian origin traveled to New Delhi, India, and acquired a urinary tract infection caused by a carbapenem-resistant Klebsiella pneumoniae strain that typed to the sequence type 14 complex. The isolate, Klebsiella pneumoniae 05-506, was shown to possess a metallo-beta-lactamase (MBL) but was negative for previously known MBL genes. Gene libraries and amplification of class 1 integrons revealed three resistance-conferring regions; the first contained bla(CMY-4) flanked by ISEcP1 and blc. The second region of 4.8 kb contained a complex class 1 integron with the gene cassettes arr-2, a new erythromycin esterase gene; ereC; aadA1; and cmlA7. An intact ISCR1 element was shown to be downstream from the qac/sul genes. The third region consisted of a new MBL gene, designated bla(NDM-1), flanked on one side by K. pneumoniae DNA and a truncated IS26 element on its other side. The last two regions lie adjacent to one another, and all three regions are found on a 180-kb region that is easily transferable to recipient strains and that confers resistance to all antibiotics except fluoroquinolones and colistin. NDM-1 shares very little identity with other MBLs, with the most similar MBLs being VIM-1/VIM-2, with which it has only 32.4% identity. As well as possessing unique residues near the active site, NDM-1 also has an additional insert between positions 162 and 166 not present in other MBLs. NDM-1 has a molecular mass of 28 kDa, is monomeric, and can hydrolyze all beta-lactams except aztreonam. Compared to VIM-2, NDM-1 displays tighter binding to most cephalosporins, in particular, cefuroxime, cefotaxime, and cephalothin (cefalotin), and also to the penicillins. NDM-1 does not bind to the carbapenems as tightly as IMP-1 or VIM-2 and turns over the carbapenems at a rate similar to that of VIM-2. In addition to K. pneumoniae 05-506, bla(NDM-1) was found on a 140-kb plasmid in an Escherichia coli strain isolated from the patient's feces, inferring the possibility of in vivo conjugation. The broad resistance carried on these plasmids is a further worrying development for India, which already has high levels of antibiotic resistance.
Figures
FIG. 1.
Alignment of novel erythromycin esterase EreC (this study) with EreA and EreA2. Amino acid changes between EreC and EreA are indicated with asterisks. The single amino acid difference between EreA and EreA2 is highlighted in gray.
FIG. 2.
Three characterized antibiotic resistance-conferring regions from K. pneumoniae 05-506. (A) The 4.3-kb region is linked to the 4.8-kb complex class 1 integron region. The genes encoding the efflux pump and lactate dehydrogenase (gray diagonal lines) are of Klebsiella origin. bla_NDM-1 (dark gray) is flanked between the pathogenicity island (vertical black lines) and IS_26/Tn_3_ (black small squares). This region lies downstream of the 4.8-kb complex class 1 integron containing Int (checkered area), arr_-2, ere2A, aadA1, and cmlA7 as gene cassettes and qac_Δ_1 (white boxes). Downstream is an intact copy of IS_CR1 (black and white diagonal lines). Arrows, direction of transcription; black ellipses, 59-base elements; Δ, genes that are truncated. (B) bla_CMY-4 (gray) is located between IS_EcP1 (black) and blc (white), as reported previously (16).
FIG. 3.
Alignment of the amino acid sequences of NDM-1 with the amino acid sequences of IMP-1, IMP-2, IMP-8, VIM-1, VIM-2, GIM-1, SPM-1, SIM-1, and KHM-1. Conserved residues coordinating zinc ions are denoted with asterisks. Key amino acids are also numbered according to the BBL scheme (12). Additional amino acids unique to NDM-1 are enclosed in a light gray box. Amino acid substitutions near the active site are highlighted in plain boxes (13).
FIG. 4.
Phylogenetic tree obtained for MBL subgroup B1. The numbers for the substitutions should be multiplied by 100. The alignment used was prepared with the CLUSTALW program. The GenBank accession numbers for the sequences are as follows: IMP-1, S71932; IMP-2, AJ243491; IMP-8, AF322577; VIM-1, Y18050; VIM-2, AF191564; GIM-1, AJ620678; SPM-1, AJ492820; SIM-1, AY887066; KHM-1, AB364006.
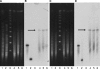
FIG. 5.
PFGE of plasmids from K. pneumoniae 05-506, conjugant E. coli J53(pNDM-1), and E. coli NF-NDM-1 (from the patient's normal flora). The bands with white arrows showed positive signals by Southern blot hybridization with the NDM-1 probe (data nor shown). K. pneumoniae 05-506 carries _bla_NDM-1 on a 180-kb plasmid, whereas E. coli NF-NDM-1 carries _bla_NDM-1 on a plasmid of approximately 140 kb.
FIG. 6.
PFGE findings (A and C) and Southern blot hybridization (B and D) with bla_NDM-1 (A and B) and an IS_CR1 probe (C and D). The genomic DNA of the index strain (K. pneumoniae 05-506) was undigested (lane 1) or was digested with S1 nuclease (lane 2), XbaI (lane 4), SpeI (lane 5), or I_-Ceu_I (lane 6). Lane 3, DNA size marker.
Similar articles
- Dissemination and Stability of the _bla_NDM-5-Carrying IncX3-Type Plasmid among Multiclonal Klebsiella pneumoniae Isolates.
Zhu W, Wang X, Qin J, Liang W, Shen Z. Zhu W, et al. mSphere. 2020 Nov 4;5(6):e00917-20. doi: 10.1128/mSphere.00917-20. mSphere. 2020. PMID: 33148824 Free PMC article. - Electron microscopic structures, serum resistance, and plasmid restructuring of New Delhi metallo-β-lactamase-1 (NDM-1)-producing ST42 Klebsiella pneumoniae emerging in Japan.
Yamamoto T, Takano T, Fusegawa T, Shibuya T, Hung WC, Higuchi W, Iwao Y, Khokhlova O, Reva I. Yamamoto T, et al. J Infect Chemother. 2013 Feb;19(1):118-27. doi: 10.1007/s10156-012-0470-z. Epub 2012 Sep 14. J Infect Chemother. 2013. PMID: 22971935 - Detection of the new metallo-beta-lactamase VIM-19 along with KPC-2, CMY-2 and CTX-M-15 in Klebsiella pneumoniae.
Pournaras S, Poulou A, Voulgari E, Vrioni G, Kristo I, Tsakris A. Pournaras S, et al. J Antimicrob Chemother. 2010 Aug;65(8):1604-7. doi: 10.1093/jac/dkq190. Epub 2010 Jun 3. J Antimicrob Chemother. 2010. PMID: 20522444 - The present danger of New Delhi metallo-β-lactamase: a threat to public health.
Qamar MU, Lopes BS, Hassan B, Khurshid M, Shafique M, Atif Nisar M, Mohsin M, Nawaz Z, Muzammil S, Aslam B, Ejaz H, Toleman MA. Qamar MU, et al. Future Microbiol. 2020 Dec;15(18):1759-1778. doi: 10.2217/fmb-2020-0069. Epub 2021 Jan 6. Future Microbiol. 2020. PMID: 33404261 Review. - Small-molecule inhibitors of bacterial-producing metallo-β-lactamases: insights into their resistance mechanisms and biochemical analyses of their activities.
Ayipo YO, Chong CF, Mordi MN. Ayipo YO, et al. RSC Med Chem. 2023 Mar 31;14(6):1012-1048. doi: 10.1039/d3md00036b. eCollection 2023 Jun 22. RSC Med Chem. 2023. PMID: 37360393 Free PMC article. Review.
Cited by
- Computationally designed peptide macrocycle inhibitors of New Delhi metallo-β-lactamase 1.
Mulligan VK, Workman S, Sun T, Rettie S, Li X, Worrall LJ, Craven TW, King DT, Hosseinzadeh P, Watkins AM, Renfrew PD, Guffy S, Labonte JW, Moretti R, Bonneau R, Strynadka NCJ, Baker D. Mulligan VK, et al. Proc Natl Acad Sci U S A. 2021 Mar 23;118(12):e2012800118. doi: 10.1073/pnas.2012800118. Proc Natl Acad Sci U S A. 2021. PMID: 33723038 Free PMC article. - NDM-4 metallo-β-lactamase with increased carbapenemase activity from Escherichia coli.
Nordmann P, Boulanger AE, Poirel L. Nordmann P, et al. Antimicrob Agents Chemother. 2012 Apr;56(4):2184-6. doi: 10.1128/AAC.05961-11. Epub 2012 Jan 17. Antimicrob Agents Chemother. 2012. PMID: 22252797 Free PMC article. - Molecular Mechanisms, Epidemiology, and Clinical Importance of β-Lactam Resistance in Enterobacteriaceae.
De Angelis G, Del Giacomo P, Posteraro B, Sanguinetti M, Tumbarello M. De Angelis G, et al. Int J Mol Sci. 2020 Jul 18;21(14):5090. doi: 10.3390/ijms21145090. Int J Mol Sci. 2020. PMID: 32708513 Free PMC article. Review. - The comprehensive antibiotic resistance database.
McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJ, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. McArthur AG, et al. Antimicrob Agents Chemother. 2013 Jul;57(7):3348-57. doi: 10.1128/AAC.00419-13. Epub 2013 May 6. Antimicrob Agents Chemother. 2013. PMID: 23650175 Free PMC article. - Emergence of a NDM-1-producing ST25 Klebsiella pneumoniae strain causing neonatal sepsis in China.
Zhao J, Zheng B, Xu H, Li J, Sun T, Jiang X, Liu W. Zhao J, et al. Front Microbiol. 2022 Oct 20;13:980191. doi: 10.3389/fmicb.2022.980191. eCollection 2022. Front Microbiol. 2022. PMID: 36338063 Free PMC article.
References
- Crowder, M. W., J. Spencer, and A. J. Vila. 2006. Metallo-β-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc. Chem. Res. 39:721-728. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous