Spatially uniform relieff (SURF) for computationally-efficient filtering of gene-gene interactions - PubMed (original) (raw)
Spatially uniform relieff (SURF) for computationally-efficient filtering of gene-gene interactions
Casey S Greene et al. BioData Min. 2009.
Abstract
Background: Genome-wide association studies are becoming the de facto standard in the genetic analysis of common human diseases. Given the complexity and robustness of biological networks such diseases are unlikely to be the result of single points of failure but instead likely arise from the joint failure of two or more interacting components. The hope in genome-wide screens is that these points of failure can be linked to single nucleotide polymorphisms (SNPs) which confer disease susceptibility. Detecting interacting variants that lead to disease in the absence of single-gene effects is difficult however, and methods to exhaustively analyze sets of these variants for interactions are combinatorial in nature thus making them computationally infeasible. Efficient algorithms which can detect interacting SNPs are needed. ReliefF is one such promising algorithm, although it has low success rate for noisy datasets when the interaction effect is small. ReliefF has been paired with an iterative approach, Tuned ReliefF (TuRF), which improves the estimation of weights in noisy data but does not fundamentally change the underlying ReliefF algorithm. To improve the sensitivity of studies using these methods to detect small effects we introduce Spatially Uniform ReliefF (SURF).
Results: SURF's ability to detect interactions in this domain is significantly greater than that of ReliefF. Similarly SURF, in combination with the TuRF strategy significantly outperforms TuRF alone for SNP selection under an epistasis model. It is important to note that this success rate increase does not require an increase in algorithmic complexity and allows for increased success rate, even with the removal of a nuisance parameter from the algorithm.
Conclusion: Researchers performing genetic association studies and aiming to discover gene-gene interactions associated with increased disease susceptibility should use SURF in place of ReliefF. For instance, SURF should be used instead of ReliefF to filter a dataset before an exhaustive MDR analysis. This change increases the ability of a study to detect gene-gene interactions. The SURF algorithm is implemented in the open source Multifactor Dimensionality Reduction (MDR) software package available from http://www.epistasis.org.
Figures
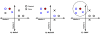
Figure 1
How Relief, ReliefF and SURF select neighbors. Each panel in this figure shows the genotypes at two markers for a dataset of cases and controls. For the purpose of this example only these two markers will be considered and both are continuous. When analyzing real data, the process of selecting neighbors is the same, however, but there will be thousands of discrete valued markers (SNPs) each of which would be represented by one of thousands of dimensions. The individual for whom neighbors are being found is shown by the filled red circle. The neighbors that each approach uses for weighting are highlighted in blue. Parts A, B, and C represent how Relief, ReliefF and SURF would select neighbors to be used in weighting. Relief selects the nearest individual of the same class (blue circle) and the nearest individual of the other class (blue cross). ReliefF selects some user specified number of individuals (two in this example) to be used for weighting. SURF, instead of using a fixed number of neighbors, uses all individuals within a distance threshold. The dotted line shows a hypothetical distance threshold.
Figure 2
Example Success Rate and Significance of Differences. Part A shows the detailed success rate analysis results for a single heritability (0.1) and sample size (1600). The success rate to filter both relevant SNPs into percentiles from the 99_th_ to 50_th_ is shown. The 99 th_percentile corresponds to the top 10 SNPs by the assigned weights in these datasets which contain 1000 SNPs. In part B pairwise comparisons are made between each pair of methods at the 99_th, 95_th_, and 75_th_ percentiles. ReliefF, SURF, TuRF, and SURF&TuRF are labeled R, S, T, and ST respectively. Significance is illustrated with levels of grey (i.e. light grey indicates 0.01 <p ≤ 0.05, dark grey indicates 0.001 <p ≤ 0.01, and black indicates p ≤ 0.001). As an example, at the 99_th_ percentile the blank square at the intersection of R and S indicates that the difference between ReliefF and SURF was not significant. On the other hand the black square at the intersection of S and ST indicates that the difference between the success rates of SURF and SURF&TuRF at that percentile was highly significant.
Figure 3
Success Rate Analysis. This is a summary of success rate as shown in figure 2 across a wide range of sample sizes and heritabilities. Within each heritability the success rates for all five genetic models for that heritability are averaged. The x_-axis for each plot corresponds to the percentiles as in figure 2. Across these situations, SuRF alone performs as well as TuRF when filtering to the 75_th percentile of SNPs. SURF outperforms ReliefF, the tuned approaches outperform the non-tuned approaches when using a more stringent filter (i.e. 99_th_ and 95_th_ percentiles), and SURF & TuRF outperforms TuRF with ReliefF.
Similar articles
- Gene-gene interaction filtering with ensemble of filters.
Yang P, Ho JW, Yang YH, Zhou BB. Yang P, et al. BMC Bioinformatics. 2011 Feb 15;12 Suppl 1(Suppl 1):S10. doi: 10.1186/1471-2105-12-S1-S10. BMC Bioinformatics. 2011. PMID: 21342539 Free PMC article. - Grid-based stochastic search for hierarchical gene-gene interactions in population-based genetic studies of common human diseases.
Moore JH, Andrews PC, Olson RS, Carlson SE, Larock CR, Bulhoes MJ, O'Connor JP, Greytak EM, Armentrout SL. Moore JH, et al. BioData Min. 2017 May 30;10:19. doi: 10.1186/s13040-017-0139-3. eCollection 2017. BioData Min. 2017. PMID: 28572842 Free PMC article. - Enabling personal genomics with an explicit test of epistasis.
Greene CS, Himmelstein DS, Nelson HH, Kelsey KT, Williams SM, Andrew AS, Karagas MR, Moore JH. Greene CS, et al. Pac Symp Biocomput. 2010:327-36. doi: 10.1142/9789814295291_0035. Pac Symp Biocomput. 2010. PMID: 19908385 Free PMC article. - [Current status of studies on genome-wide gene-gene interactions].
Shen JW, Hu XH, Shi YY. Shen JW, et al. Yi Chuan. 2011 Aug;33(8):820-8. doi: 10.3724/sp.j.1005.2011.00820. Yi Chuan. 2011. PMID: 21831799 Review. Chinese. - Relief-based feature selection: Introduction and review.
Urbanowicz RJ, Meeker M, La Cava W, Olson RS, Moore JH. Urbanowicz RJ, et al. J Biomed Inform. 2018 Sep;85:189-203. doi: 10.1016/j.jbi.2018.07.014. Epub 2018 Jul 18. J Biomed Inform. 2018. PMID: 30031057 Free PMC article. Review.
Cited by
- HSD3B and gene-gene interactions in a pathway-based analysis of genetic susceptibility to bladder cancer.
Andrew AS, Hu T, Gu J, Gui J, Ye Y, Marsit CJ, Kelsey KT, Schned AR, Tanyos SA, Pendleton EM, Mason RA, Morlock EV, Zens MS, Li Z, Moore JH, Wu X, Karagas MR. Andrew AS, et al. PLoS One. 2012;7(12):e51301. doi: 10.1371/journal.pone.0051301. Epub 2012 Dec 19. PLoS One. 2012. PMID: 23284679 Free PMC article. - Assessing the limitations of relief-based algorithms in detecting higher-order interactions.
Freda PJ, Ye S, Zhang R, Moore JH, Urbanowicz RJ. Freda PJ, et al. BioData Min. 2024 Oct 1;17(1):37. doi: 10.1186/s13040-024-00390-0. BioData Min. 2024. PMID: 39354639 Free PMC article. - Transition-transversion encoding and genetic relationship metric in ReliefF feature selection improves pathway enrichment in GWAS.
Arabnejad M, Dawkins BA, Bush WS, White BC, Harkness AR, McKinney BA. Arabnejad M, et al. BioData Min. 2018 Nov 3;11:23. doi: 10.1186/s13040-018-0186-4. eCollection 2018. BioData Min. 2018. PMID: 30410580 Free PMC article. - ExSTraCS 2.0: Description and Evaluation of a Scalable Learning Classifier System.
Urbanowicz RJ, Moore JH. Urbanowicz RJ, et al. Evol Intell. 2015 Sep;8(2):89-116. doi: 10.1007/s12065-015-0128-8. Epub 2015 Apr 3. Evol Intell. 2015. PMID: 26417393 Free PMC article. - Application of a spatially-weighted Relief algorithm for ranking genetic predictors of disease.
Stokes ME, Visweswaran S. Stokes ME, et al. BioData Min. 2012 Dec 3;5(1):20. doi: 10.1186/1756-0381-5-20. BioData Min. 2012. PMID: 23198930 Free PMC article.
References
Grants and funding
- P42 ES007373/ES/NIEHS NIH HHS/United States
- R01 AI059694/AI/NIAID NIH HHS/United States
- R01 HD047447/HD/NICHD NIH HHS/United States
- R01 LM009012/LM/NLM NIH HHS/United States
LinkOut - more resources
Full Text Sources