Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast - PubMed (original) (raw)
Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast
Helen E M Stimpson et al. Mol Biol Cell. 2009 Nov.
Abstract
Recent studies have revealed the detailed timing of protein recruitment to endocytic sites in budding yeast. However, little is understood about the early stages of their formation. Here we identify the septin-associated protein Syp1p as a component of the machinery that drives clathrin-mediated endocytosis in budding yeast. Syp1p arrives at endocytic sites early in their formation and shares unique dynamics with the EH-domain protein Ede1p. We find that Syp1p is related in amino acid sequence to several mammalian proteins one of which, SGIP1-alpha, is an endocytic component that binds the Ede1p homolog Eps15. Like Syp1p, SGIP1-alpha arrives early at sites of clathrin-mediated endocytosis, suggesting that Syp1p/Ede1p and SGIP1-alpha/Eps15 may have a conserved function. In yeast, both Syp1p and Ede1p play important roles in the rate of endocytic site turnover. Additionally, Ede1p is important for endocytic site formation, whereas Syp1p acts as a polarized factor that recruits both Ede1p and endocytic sites to the necks of emerging buds. Thus Ede1p and Syp1p are conserved, early-arriving endocytic proteins with roles in the formation and placement of endocytic sites, respectively.
Figures
Figure 1.
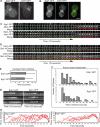
Syp1p and Ede1p dynamics at endocytic sites. (A) Epifluorescence images of live wild-type cells expressing Syp1-GFP or Ede1-GFP (strains DDY3865 and -3866). (B) Epifluorescence images of a single cell coexpressing Syp1-GFP and Ede1-RFP (strain DDY3871). (C) Dynamic cell surface localization of Syp1-GFP and Ede1-GFP (viewed by TIRF microscopy) in reference to Abp1-RFP (viewed by epifluorescence microscopy). Time series shows individual patches from two-color movies. Frame interval, 2 s. The proteins were coexpressed in strains DDY3867 and -3868. (D) Dynamic cell surface localization of Syp1-GFP and Ede1-GFP (viewed by TIRF microscopy) in reference to Sla1-mCherry (viewed by epifluorescence microscopy). Time series shows individual patches from two-color movies. Frame interval, 2 s. The proteins were coexpressed in DDY3899 and -3900. (E) Average lifetimes of Syp1-GFP and Ede1-GFP patches ± SD, n = 60. Data taken from 4-min TIRF microscopy movies with 2-s frame intervals. (F) Distribution of Syp1-GFP and Ede1-GFP patch lifetimes from the same dataset as in C. (G) Kymographs of representative Syp1-GFP and Ede1-GFP patches from epifluorescence movies. Patches were oriented so the outside of the cell is on top. (H) Quantification of fluorescence intensity for individual Syp1-GFP and Ede1-GFP patches over time. Each curve represents data from one patch. Fluorescence intensity was corrected for photobleaching. Movies were taken with 2-s frame intervals. All scale bars, 2 μm.
Figure 2.
SGIP1-α is a Syp1p homolog. (A) Sequence alignment of Syp1p, SGIP1-α (Mus musculus, NP_659155.1), FCHo1 (H. sapiens, NP_055937.1), and FCHo2 (H. sapiens, NP_620137). Key to alignment: red, complete consensus; blue, consensus of two or three sequences; #, conserved as D/E.; and !, conserved as I/V. (B) Predicted domain structures of Syp1p, SGIP1-α, FCHo1, and FCHo2. MP, membrane phospholipid-binding domain; SAFF, a PFAM predicted domain. (C) Colocalization of SGIP1-α-GFP and DsRed-clathrin (mouse clathrin light chain a) at the cell surface by TIRF microscopy in Swiss 3T3 cells. Scale bar, 2 μm. (D) Dynamic cell surface localization of SGIP1-α-GFP and DsRed-clathrin by TIRF microscopy. Three representative endocytic site assembly (top) and disassembly (bottom) events are shown. For each event, a montage of DsRed-clathrin and SGIP1-α-GFP recruitment to endocytic sites is shown (frame interval, 12 s), as well as quantification of fluorescence intensity over time (frame interval, 2 s). The region of the graph corresponding to the montage is outlined in gray.
Figure 3.
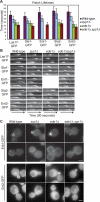
The endocytic role of Syp1p and Ede1p. (A) Patch lifetimes of Las17-GFP, Sla1-GFP, Sla2-GFP, Ent1-GFP, and Ent2-GFP in wild-type, _syp1_Δ, _ede1_Δ, and _syp1_Δ _ede1_Δ cells (strains DDY2736, -3696, -3697, -3700, -3701, -3798, and -3872–3885). n = 30 patches for each strain. Error bars, SD. Frame rate was 1 frame per second. (B) Kymographs of representative Las17-GFP, Sla1-GFP, Sla2-GFP, Ent1-GFP, and Ent2-GFP patches from epifluorescence movies of wild-type, _syp1_Δ, _ede1_Δ, and _syp1_Δ _ede1_Δ cells (the same strains listed in A). Patches are oriented so the outside of the cell is on top. (C) Epifluorescence images of live wild-type _syp1_Δ, _ede1_Δ, and _syp1_Δ _ede1_Δ cells expressing Ent1-GFP or Ent2-GFP (strains DDY3696, -3697, and -3880–3885). Top, small-budded cells; bottom, large-budded cells. Scale bars, 2 μm.
Figure 4.
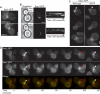
Localization interdependence of Syp1p and Ede1p. (A) Epifluorescence images of wild-type and _ede1_Δ cells expressing Syp1-GFP (strains DDY3865 and -3869). (B) Dynamic surface localization of Syp1-GFP in wild-type and _ede1_Δ cells viewed by TIRF microscopy. Left, single frames from movies; Right, kymograph representations of the same movies. Movies were taken with 1-s frame intervals for 2 min. (C) Epifluorescence images of small- (top), medium- (middle), and large-budded (bottom) wild-type and _syp1_Δ cells expressing Ede1-GFP (strains DDY3866 and -3870). Arrowheads, the bud neck region affected by SYP1 deletion. (D) Cell cycle distribution of Syp1-GFP and Ede1-RFP in wild-type cells (strain DDY3871). Individual cells were imaged every 10 min for 2 h, and one image is shown for every 20 min (the full sequence can be seen in Supplemental Movie S2); green and red images were acquired immediately after one another. White arrowheads, the first signs of neck localization during bud emergence (a second cell cycle has begun at 120 min); red arrowheads, localization to the bud neck at cytokinesis. Scale bars, 2 μm throughout.
Figure 5.
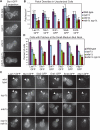
The early module regulates endocytic site formation and distribution. (A) Maximum intensity projections of Z-stacks of wild-type, _syp1_Δ, _ede1_Δ, and _syp1_Δ _ede1_Δ cells expressing Sla1-GFP (strains DDY3700, -3798, -3875, and -3876). Z-stacks were acquired through the entire cell at 0.15-μm intervals. Scale bar, 2 μm. (B) Patch number per cell surface area (μm2) in wild-type, _syp1_Δ, _ede1_Δ, and _syp1_Δ _ede1_Δ cells expressing the indicated GFP-tagged markers (strains DDY2736, -3696, -3697, -3700, -3701, -3798, and -3872–3885). n = 20 cells for each strain; error bars, SD. Patches were counted in approximately spherical unbudded or large-budded cells; cell surface area was estimated as an average of sphere surface areas calculated from four diameters measured from the maximum intensity projections. (C) Maximum intensity projections of Z-stacks of small-budded wild-type, _syp1_Δ, _ede1_Δ, and _syp1_Δ _ede1_Δ cells expressing the indicated GFP-tagged markers (strains DDY2736, -3696, -3697, -3700, -3701, -3798, and -3872–3889). Z-stacks were acquired through the entire cell with 0.15-μm intervals. Arrowheads indicate the bud neck region affected by SYP1 deletion. Scale bar, 2 μm. (D) Percentage of wild-type, _syp1_Δ, _ede1_Δ, and _syp1_Δ _ede1_Δ cells expressing the indicated GFP-tagged markers with patches polarized to the small-budded neck (using the same strains listed in C). Mean averages from three separate experiments are shown (n = 60 cells for each strain in each experiment; error bars, SD). Cells were scored as described in the text.
Figure 6.
Model for the role of the early module in endocytic site formation. (A) Temporal relationship of the endocytic modules including the newly defined early module. (B) Roles and interactions for the early module in endocytic site organization. In our model, Ede1p and Syp1p interact with the endocytic and polarity machinery, respectively, to initiate endocytic site formation and control spatial distribution.
Comment in
- Mol Biol Cell. 20:4629.
Similar articles
- Analysis of yeast endocytic site formation and maturation through a regulatory transition point.
Carroll SY, Stimpson HE, Weinberg J, Toret CP, Sun Y, Drubin DG. Carroll SY, et al. Mol Biol Cell. 2012 Feb;23(4):657-68. doi: 10.1091/mbc.E11-02-0108. Epub 2011 Dec 21. Mol Biol Cell. 2012. PMID: 22190733 Free PMC article. - Regulation of clathrin coat assembly by Eps15 homology domain-mediated interactions during endocytosis.
Suzuki R, Toshima JY, Toshima J. Suzuki R, et al. Mol Biol Cell. 2012 Feb;23(4):687-700. doi: 10.1091/mbc.E11-04-0380. Epub 2011 Dec 21. Mol Biol Cell. 2012. PMID: 22190739 Free PMC article. - A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis.
Gagny B, Wiederkehr A, Dumoulin P, Winsor B, Riezman H, Haguenauer-Tsapis R. Gagny B, et al. J Cell Sci. 2000 Sep;113 ( Pt 18):3309-19. doi: 10.1242/jcs.113.18.3309. J Cell Sci. 2000. PMID: 10954428 - Function and regulation of Saccharomyces cerevisiae myosins-I in endocytic budding.
Giblin J, Fernández-Golbano IM, Idrissi FZ, Geli MI. Giblin J, et al. Biochem Soc Trans. 2011 Oct;39(5):1185-90. doi: 10.1042/BST0391185. Biochem Soc Trans. 2011. PMID: 21936786 Review. - Taking apart the endocytic machinery.
Kaksonen M. Kaksonen M. J Cell Biol. 2008 Mar 24;180(6):1059-60. doi: 10.1083/jcb.200802174. J Cell Biol. 2008. PMID: 18362177 Free PMC article. Review.
Cited by
- Condensation of Ede1 promotes the initiation of endocytosis.
Kozak M, Kaksonen M. Kozak M, et al. Elife. 2022 Apr 12;11:e72865. doi: 10.7554/eLife.72865. Elife. 2022. PMID: 35412456 Free PMC article. - Lessons from yeast for clathrin-mediated endocytosis.
Boettner DR, Chi RJ, Lemmon SK. Boettner DR, et al. Nat Cell Biol. 2011 Dec 22;14(1):2-10. doi: 10.1038/ncb2403. Nat Cell Biol. 2011. PMID: 22193158 Free PMC article. Review. - Role of Clathrin and Dynamin in Clathrin Mediated Endocytosis/Synaptic Vesicle Recycling and Implications in Neurological Diseases.
Prichard KL, O'Brien NS, Murcia SR, Baker JR, McCluskey A. Prichard KL, et al. Front Cell Neurosci. 2022 Jan 18;15:754110. doi: 10.3389/fncel.2021.754110. eCollection 2021. Front Cell Neurosci. 2022. PMID: 35115907 Free PMC article. Review. - Candida albicans END3 Mediates Endocytosis and Has Subsequent Roles in Cell Wall Integrity, Morphological Switching, and Tissue Invasion.
Rollenhagen C, Agyeman H, Eszterhas S, Lee SA. Rollenhagen C, et al. Microbiol Spectr. 2022 Apr 27;10(2):e0188021. doi: 10.1128/spectrum.01880-21. Epub 2022 Mar 2. Microbiol Spectr. 2022. PMID: 35234488 Free PMC article. - A Flow Cytometry-Based Phenotypic Screen To Identify Novel Endocytic Factors in Saccharomyces cerevisiae.
Wrasman K, Alioto SL, Zhang Y, Hoban K, Khairy M, Goode BL, Wendland B. Wrasman K, et al. G3 (Bethesda). 2018 May 4;8(5):1497-1512. doi: 10.1534/g3.118.200102. G3 (Bethesda). 2018. PMID: 29540444 Free PMC article.
References
- Aguilar R. C., Watson H. A., Wendland B. The yeast Epsin Ent1 is recruited to membranes through multiple independent interactions. J. Biol. Chem. 2003;278:10737–10743. - PubMed
- Collins S. R., Kemmeren P., Zhao X. C., Greenblatt J. F., Spencer F., Holstege F. C., Weissman J. S., Krogan N. J. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell Proteom. 2007;6:439–450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM065462/GM/NIGMS NIH HHS/United States
- R35 GM118149/GM/NIGMS NIH HHS/United States
- GM50399/GM/NIGMS NIH HHS/United States
- R01 GM050399/GM/NIGMS NIH HHS/United States
- GM65462/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous