Reconstructing Indian population history - PubMed (original) (raw)
Reconstructing Indian population history
David Reich et al. Nature. 2009.
Abstract
India has been underrepresented in genome-wide surveys of human variation. We analyse 25 diverse groups in India to provide strong evidence for two ancient populations, genetically divergent, that are ancestral to most Indians today. One, the 'Ancestral North Indians' (ANI), is genetically close to Middle Easterners, Central Asians, and Europeans, whereas the other, the 'Ancestral South Indians' (ASI), is as distinct from ANI and East Asians as they are from each other. By introducing methods that can estimate ancestry without accurate ancestral populations, we show that ANI ancestry ranges from 39-71% in most Indian groups, and is higher in traditionally upper caste and Indo-European speakers. Groups with only ASI ancestry may no longer exist in mainland India. However, the indigenous Andaman Islanders are unique in being ASI-related groups without ANI ancestry. Allele frequency differences between groups in India are larger than in Europe, reflecting strong founder effects whose signatures have been maintained for thousands of years owing to endogamy. We therefore predict that there will be an excess of recessive diseases in India, which should be possible to screen and map genetically.
Figures
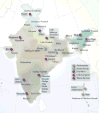
Figure 1
Map of India with the state of origin of the 25 groups that we studied.
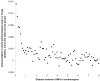
Figure 2
Linkage disequilibrium based evidence for founder events in India. For each pair of samples, we calculate the autocorrelation of the number of shared alleles as a function of distance, recognizing that SNP genotypes should differ by at most one allele in regions of identity by descent. To correct for background allele sharing, we subtract the same quantity comparing across groups. Allele sharing in the Vysya decreases with an exponential decay of 0.461 cM as shown here, suggesting a founder event roughly 100/(2*0.461) = 108 generations ago. We present similar analyses for all Indian groups in Figure S2.
Figure 3
Principal components analysis (PCA) of 22 groups from the Indian subcontinent. Analysis of these groups along with Europeans (CEU) and Chinese (CHB) reveals a gradient of relatedness to CEU that runs through the majority of Indo-European and Dravidian groups, with the Kashmiri Pandit most related to CEU. Both the Austro-Asiatic speaking groups (Kharia and Santhal) and the tribal Sahariya are off-cline, while the two Tibeto-Burman speaking groups cluster with CHB. (Data from the outlying Siddi, Onge and Great Andamanese are not shown.)
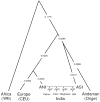
Figure 4
A model relating the history of Indian and non-Indian groups. Modeling the Pathan, Vaish, Meghawal and Bhil as mixtures of ANI and ASI, and relating them to non-Indians by the phylogenetic tree (YRI,(CEU,ANI),(ASI, Onge))), provides an excellent fit to the data. While the model is precise about tree topology and ordering of splits, it provides no information about population size changes or the timings of events. We estimate genetic drift on each lineage in the sense of variance in allele frequencies, which we rescale to be comparable to FST (standard errors are typically ±0.001 but are not shown).
Comment in
- Human genetics: Tracing India's invisible threads.
Chakravarti A. Chakravarti A. Nature. 2009 Sep 24;461(7263):487-8. doi: 10.1038/461487a. Nature. 2009. PMID: 19779444 No abstract available.
Similar articles
- Genetic affinities among the lower castes and tribal groups of India: inference from Y chromosome and mitochondrial DNA.
Thanseem I, Thangaraj K, Chaubey G, Singh VK, Bhaskar LV, Reddy BM, Reddy AG, Singh L. Thanseem I, et al. BMC Genet. 2006 Aug 7;7:42. doi: 10.1186/1471-2156-7-42. BMC Genet. 2006. PMID: 16893451 Free PMC article. - Genetic evidence for recent population mixture in India.
Moorjani P, Thangaraj K, Patterson N, Lipson M, Loh PR, Govindaraj P, Berger B, Reich D, Singh L. Moorjani P, et al. Am J Hum Genet. 2013 Sep 5;93(3):422-38. doi: 10.1016/j.ajhg.2013.07.006. Epub 2013 Aug 8. Am J Hum Genet. 2013. PMID: 23932107 Free PMC article. - Presence of three different paternal lineages among North Indians: a study of 560 Y chromosomes.
Zhao Z, Khan F, Borkar M, Herrera R, Agrawal S. Zhao Z, et al. Ann Hum Biol. 2009 Jan-Feb;36(1):46-59. doi: 10.1080/03014460802558522. Ann Hum Biol. 2009. PMID: 19058044 Free PMC article. - Genetic evidence on the origins of Indian caste populations.
Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BV, Reddy PG, Rasanayagam A, Papiha SS, Villems R, Redd AJ, Hammer MF, Nguyen SV, Carroll ML, Batzer MA, Jorde LB. Bamshad M, et al. Genome Res. 2001 Jun;11(6):994-1004. doi: 10.1101/gr.gr-1733rr. Genome Res. 2001. PMID: 11381027 Free PMC article. - The genetic makings of South Asia.
Metspalu M, Mondal M, Chaubey G. Metspalu M, et al. Curr Opin Genet Dev. 2018 Dec;53:128-133. doi: 10.1016/j.gde.2018.09.003. Epub 2018 Oct 1. Curr Opin Genet Dev. 2018. PMID: 30286387 Review.
Cited by
- North African populations carry the signature of admixture with Neandertals.
Sánchez-Quinto F, Botigué LR, Civit S, Arenas C, Avila-Arcos MC, Bustamante CD, Comas D, Lalueza-Fox C. Sánchez-Quinto F, et al. PLoS One. 2012;7(10):e47765. doi: 10.1371/journal.pone.0047765. Epub 2012 Oct 17. PLoS One. 2012. PMID: 23082212 Free PMC article. - Indian Ocean crossroads: human genetic origin and population structure in the Maldives.
Pijpe J, de Voogt A, van Oven M, Henneman P, van der Gaag KJ, Kayser M, de Knijff P. Pijpe J, et al. Am J Phys Anthropol. 2013 May;151(1):58-67. doi: 10.1002/ajpa.22256. Epub 2013 Mar 21. Am J Phys Anthropol. 2013. PMID: 23526367 Free PMC article. - Association of TCF7L2 and ADIPOQ with body mass index, waist-hip ratio, and systolic blood pressure in an endogamous ethnic group of India.
Gupta V, Khadgawat R, Ng HK, Walia GK, Kalla L, Rao VR, Sachdeva MP. Gupta V, et al. Genet Test Mol Biomarkers. 2012 Aug;16(8):948-51. doi: 10.1089/gtmb.2012.0051. Epub 2012 May 14. Genet Test Mol Biomarkers. 2012. PMID: 22583123 Free PMC article. - EigenGWAS: finding loci under selection through genome-wide association studies of eigenvectors in structured populations.
Chen GB, Lee SH, Zhu ZX, Benyamin B, Robinson MR. Chen GB, et al. Heredity (Edinb). 2016 Jul;117(1):51-61. doi: 10.1038/hdy.2016.25. Epub 2016 May 4. Heredity (Edinb). 2016. PMID: 27142779 Free PMC article. - Recent advances in the study of fine-scale population structure in humans.
Novembre J, Peter BM. Novembre J, et al. Curr Opin Genet Dev. 2016 Dec;41:98-105. doi: 10.1016/j.gde.2016.08.007. Epub 2016 Sep 20. Curr Opin Genet Dev. 2016. PMID: 27662060 Free PMC article. Review.
References
- Majumbdar DN, Rao CR. Race elements in Bengal: A quantitative study (with a forward by P C Mahalanobis) Calcutta: Asia Publishing House and Statistical Publishing Society; 1960.
- Roychoudhury AK, Nei M. Genetic relationships between Indians and their neighboring populations. Hum Hered. 1985;35:201–206. - PubMed
- Das BM, Das PB, Das R, Walter H, Danker-Hopfe H. Anthropological studies in Assam, India. Anthropologischer Anzeiger. 1986;44:239–248. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials