TbPIF5 is a Trypanosoma brucei mitochondrial DNA helicase involved in processing of minicircle Okazaki fragments - PubMed (original) (raw)
TbPIF5 is a Trypanosoma brucei mitochondrial DNA helicase involved in processing of minicircle Okazaki fragments
Beiyu Liu et al. PLoS Pathog. 2009 Sep.
Abstract
Trypanosoma brucei's mitochondrial genome, kinetoplast DNA (kDNA), is a giant network of catenated DNA rings. The network consists of a few thousand 1 kb minicircles and several dozen 23 kb maxicircles. Here we report that TbPIF5, one of T. brucei's six mitochondrial proteins related to Saccharomyces cerevisiae mitochondrial DNA helicase ScPIF1, is involved in minicircle lagging strand synthesis. Like its yeast homolog, TbPIF5 is a 5' to 3' DNA helicase. Together with other enzymes thought to be involved in Okazaki fragment processing, TbPIF5 localizes in vivo to the antipodal sites flanking the kDNA. Minicircles in wild type cells replicate unidirectionally as theta-structures and are unusual in that Okazaki fragments are not joined until after the progeny minicircles have segregated. We now report that overexpression of TbPIF5 causes premature removal of RNA primers and joining of Okazaki fragments on theta structures. Further elongation of the lagging strand is blocked, but the leading strand is completed and the minicircle progeny, one with a truncated H strand (ranging from 0.1 to 1 kb), are segregated. The minicircles with a truncated H strand electrophorese on an agarose gel as a smear. This replication defect is associated with kinetoplast shrinkage and eventual slowing of cell growth. We propose that TbPIF5 unwinds RNA primers after lagging strand synthesis, thus facilitating processing of Okazaki fragments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
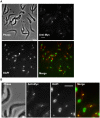
Figure 1. Localization of TbPIF5-Myc.
Procyclic 927 cells harboring c-Myc-tagged TbPIF5 were fixed with 3% paraformaldehyde and then adhered to poly-L-lysine-treated slides. Immunostaining for TbPIF5-Myc used 1∶100 rabbit anti-Myc polyclonal antibody (Santa Cruz) and 1∶600 Alexa Fluor 568-conjugated goat anti-rabbit IgG (Molecular Probes). Conditions for fixing, permeabilizing, and staining cells were described . In the merged image, anti-Myc is in red and DAPI in green. Arrows in panel A point out two cells with an enlarged magnification in panel B. Bar, 5 µm.
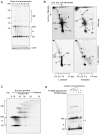
Figure 2. ATPase and helicase assays of recombinant TbPIF5.
(A) Coomassie-stained SDS-PAGE gel and Western blot of purified recombinant TbPIF5. (B) Assay of TbPIF5 ATPase activity. The substrates and products were separated by polyethyleneimine thin layer chromatography; arrow shows origin. The [32P]Pi standard in the left-hand lane was prepared from [γ-32P] ATP by boiling 5 min in 1 M HCl. (C) Assay of TbPIF5 helicase activity. Substrates and products were separated by 12% polyacrylamide gel electrophoresis. (D) Helicase activity was measured at various concentrations of Mg++-ATP. (E) Assay of polarity of TbPIF5 helicase activity. Helicase substrates are diagrammed in Panels C (strand lengths are not to scale) and E (strand lengths for oligonucleotides a, b and c are 21, 21 and 90 nucleotides). * indicates 5′ 32P end label.
Figure 3. Effects of TbPIF5 overexpression.
(A) Effect of TbPIF5 overexpression on cell growth. Overexpression was induced by addition of tetracycline (1 µg/ml) at day 0. The value of parasites/ml on the y-axis is the measured value times the dilution factor. Inset, Northern blot of mRNA level without or with overexpression. (B) Effect of overexpression on kinetoplast size as visualized by fluorescence microscopy of cells stained with DAPI (5 µg/ml). K, kinetoplast; N, nucleus. Bar, 5 µm. (C) Kinetics of kDNA loss as determined by visual analysis of images (>200 randomly-selected DAPI-stained cells for each time point). Inset images are examples of a cell with normal kinetoplast, small kinetoplast and no kinetoplast (kinetoplast is marked by arrow). (D) Effect of TbPIF5 overexpression on minicircle and maxicircle abundance. Total maxicircles (Maxi) and minicircles (Mini) were detected by probing a Southern blot after the total DNA (106 cell equivalents/lane) was digested with Hind III/XbaI and fractionated onto an agarose gel. The maxicircle probe detects only the 1.4 kb fragment, and only the 1 kb fragment derived from the heterogeneous minicircle population is shown. A hexose transporter fragment was probed as a loading control (Load). (E) Quantitation of the Southern blot in Fig. 3D showing maxicircle and minicircle species as indicated. Values represent the abundance of minicircle/maxicircle relative to its abundance in the uninduced cells. Values were normalized to load control.
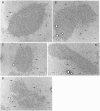
Figure 4. Electron micrographs of kDNA networks from TbPIF5 overexpression cells.
(A) and (B), kDNA isolated from wild-type cells. (C–E), kDNA isolated from TbPIF5 overexpression cells six days after induction. Arrow, maxicircle loops. Bar, 500 nm.
Figure 5. Effect of TbPIF5 overexpression on free minicircle intermediates.
(A) Total DNA (106 cell equivalents/lane) was fractionated on a 1.5% agarose gel in TBE buffer (both the gel and running buffer contained 1 µg/ml ethidium bromide). A southern blot was probed for minicircles and hexose transporter (Load). (B) Neutral/alkaline two-dimensional gel electrophoresis. Total DNA from 3×107 wild type or induced cells (1 day overexpression) was fractionated on a two-dimensional gel. Strand-specific hybridizations were conducted with synthetic oligonucleotide probes. The upper panel shows a longer exposure version of the same 2-D gel used in Fig. 3F of our previous paper . The scales below the panels indicate the sizes of linear markers in the second dimension. (C) Sedimentation of free minicircle intermediates in a 5–20% sucrose gradient. Fractions were collected from the top (1 ml fractions), subjected to electrophoresis, and assayed by probing a Southern blot . (D) Gel electrophoresis (using conditions described for Panel A) of free minicircles treated with various enzymes. Total free minicircles were purified on the sucrose gradient in Panel C by pooling fractions 8 to 16 and ethanol precipitating the DNA. The free minicircles were then treated with T4 DNA polymerase (0.6 U, 1 U, 2 U and 3 U, New England Biolabs) and/or T4 DNA ligase (400 U, New England Biolabs). CM, catenated minicircles; N/G, nicked/gapped minicircles; CC, covalently-closed minicircles; θ, theta-structure; k, knotted minicircle; H, fraction H; ccD, covalently-closed dimer; ccT, covalently-closed trimer; nD, nicked dimer; L, linearized minicircle; MG, multiply-gapped minicircle; OF, Okazaki fragments.
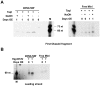
Figure 6. Effect of TbPIF5 overexpression on replication primers.
See for experimental details of this experiment. (A) Analysis of 5′ ribonucleotides on the first Okazaki fragment. kDNA networks and free minicircle intermediates were isolated from cells without TbPIF5 overexpression or after overexpression for 1 day. DNA was digested with TaqI and treated with 0.3 M NaOH as indicated; alkali treatment would remove ribonucleotides and alter fragment mobility. After fractionation on a denaturing 9% polyacrylamide gel, a Southern blot was probed for the first Okazaki fragment with a 32P-labeled oligonucleotide. (B) Analysis of primers on the leading strand. kDNA networks and free minicircle intermediates were digested with HpyCH4V, and after electrophoresis a Southern blot was probed with a 32P-labeled oligonucleotide complementary to the 5′ end of the leading strand. M, size marker; OE, overexpression.
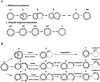
Figure 7. Comparison of the normal free minicircle replication mechanism with that in cells overexpressing TbPIF5.
(A) Replication scheme showing (line 1) conversion, via theta structures (θ), of covalently-closed parental minicircles to gapped (G) and multiply-gapped (MG) progeny. The MG molecules are then converted (line 2) to nicked minicircles (N). The thick strand is H, which is synthesized discontinuously by Okazaki fragments, and the thin strand is L, which is synthesized continuously. * is an RNA primer and horizontal lines linking two circles in the θ-structures represent base pairs in the unreplicated portion. (B) When TbPIF5 is overexpressed, it binds to θ-structures and triggers primer removal. Removal of primers between the newly-synthesized Okazaki fragments generates gaps which can then be repaired. A similar removal of primers that have not yet been used for initiation blocks subsequent lagging strand replication. Leading strand synthesis is essentially unaffected and proceeds to completion allowing segregation. This process generates fraction H, a family of minicircles with a circular parental L-strand and increasing numbers of joined Okazaki fragments (ranging in size from 73 nt, the first Okazaki fragment, to 1 kb). This diagram shows the generation of three different species of fraction H, in which one, two or three Okazaki fragments are synthesized and then subjected to primer removal and joining. For simplicity, this diagram was not drawn to scale. We speculate that reactions in line 1 of panel A occur in the KFZ and those in line 2 take place in the antipodal sites. We further speculate that all reactions in Panel B occur in the KFZ. An alternative explanation for the existence of fraction H is that TbPIF5 overexpression somehow causes failure of the coordination of leading and lagging strand replication, so that the lagging strand is now synthesized continuously. Further studies are needed to test this possibility.
Similar articles
- TbPIF1, a Trypanosoma brucei mitochondrial DNA helicase, is essential for kinetoplast minicircle replication.
Liu B, Yildirir G, Wang J, Tolun G, Griffith JD, Englund PT. Liu B, et al. J Biol Chem. 2010 Mar 5;285(10):7056-66. doi: 10.1074/jbc.M109.084038. Epub 2009 Dec 30. J Biol Chem. 2010. PMID: 20042610 Free PMC article. - Effects of RNA interference of Trypanosoma brucei structure-specific endonuclease-I on kinetoplast DNA replication.
Liu Y, Motyka SA, Englund PT. Liu Y, et al. J Biol Chem. 2005 Oct 21;280(42):35513-20. doi: 10.1074/jbc.M507296200. Epub 2005 Aug 11. J Biol Chem. 2005. PMID: 16096280 - Trypanosomes have six mitochondrial DNA helicases with one controlling kinetoplast maxicircle replication.
Liu B, Wang J, Yaffe N, Lindsay ME, Zhao Z, Zick A, Shlomai J, Englund PT. Liu B, et al. Mol Cell. 2009 Aug 28;35(4):490-501. doi: 10.1016/j.molcel.2009.07.004. Epub 2009 Jul 30. Mol Cell. 2009. PMID: 19646907 Free PMC article. - The structure and replication of kinetoplast DNA.
Shlomai J. Shlomai J. Curr Mol Med. 2004 Sep;4(6):623-47. doi: 10.2174/1566524043360096. Curr Mol Med. 2004. PMID: 15357213 Review. - The structure and replication of kinetoplast DNA.
Shapiro TA, Englund PT. Shapiro TA, et al. Annu Rev Microbiol. 1995;49:117-43. doi: 10.1146/annurev.mi.49.100195.001001. Annu Rev Microbiol. 1995. PMID: 8561456 Review.
Cited by
- A leucine aminopeptidase is involved in kinetoplast DNA segregation in Trypanosoma brucei.
Peña-Diaz P, Vancová M, Resl C, Field MC, Lukeš J. Peña-Diaz P, et al. PLoS Pathog. 2017 Apr 7;13(4):e1006310. doi: 10.1371/journal.ppat.1006310. eCollection 2017 Apr. PLoS Pathog. 2017. PMID: 28388690 Free PMC article. - Mitochondrial heat shock protein machinery hsp70/hsp40 is indispensable for proper mitochondrial DNA maintenance and replication.
Týč J, Klingbeil MM, Lukeš J. Týč J, et al. mBio. 2015 Feb 10;6(1):e02425-14. doi: 10.1128/mBio.02425-14. mBio. 2015. PMID: 25670781 Free PMC article. - Unwinding the functions of the Pif1 family helicases.
Bochman ML, Sabouri N, Zakian VA. Bochman ML, et al. DNA Repair (Amst). 2010 Mar 2;9(3):237-49. doi: 10.1016/j.dnarep.2010.01.008. Epub 2010 Jan 25. DNA Repair (Amst). 2010. PMID: 20097624 Free PMC article. Review. - A second mitochondrial DNA primase is essential for cell growth and kinetoplast minicircle DNA replication in Trypanosoma brucei.
Hines JC, Ray DS. Hines JC, et al. Eukaryot Cell. 2011 Mar;10(3):445-54. doi: 10.1128/EC.00308-10. Epub 2011 Jan 21. Eukaryot Cell. 2011. PMID: 21257796 Free PMC article. - PNT1 Is a C11 Cysteine Peptidase Essential for Replication of the Trypanosome Kinetoplast.
Grewal JS, McLuskey K, Das D, Myburgh E, Wilkes J, Brown E, Lemgruber L, Gould MK, Burchmore RJ, Coombs GH, Schnaufer A, Mottram JC. Grewal JS, et al. J Biol Chem. 2016 Apr 29;291(18):9492-500. doi: 10.1074/jbc.M116.714972. Epub 2016 Mar 3. J Biol Chem. 2016. PMID: 26940875 Free PMC article.
References
- Sogin ML, Silberman JD. Evolution of the protists and protistan parasites from the perspective of molecular systematics. Int J Parasitol. 1998;28:11–20. - PubMed
- Liu B, Liu Y, Motyka SA, Agbo EE, Englund PT. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 2005;21:363–369. - PubMed
- Shlomai J. The structure and replication of kinetoplast DNA. Curr Mol Med. 2004;4:623–647. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases