Mutational analysis of Drosophila basigin function in the visual system - PubMed (original) (raw)
Mutational analysis of Drosophila basigin function in the visual system
Michelle Munro et al. Gene. 2010.
Abstract
Drosophila basigin is a cell-surface glycoprotein of the Ig superfamily and a member of a protein family that includes mammalian EMMPRIN/CD147/basigin, neuroplastin, and embigin. Our previous work on Drosophila basigin has shown that it is required for normal photoreceptor cell structure and normal neuron-glia interaction in the fly visual system. Specifically, the photoreceptor neurons of mosaic animals that are mutant in the eye for basigin show altered cell structure with nuclei, mitochondria and rER misplaced and variable axon diameter compared to wild-type. In addition, glia cells in the optic lamina that contact photoreceptor axons are misplaced and show altered structure. All these defects are rescued by expression of either transgenic fly basigin or transgenic mouse basigin in the photoreceptors demonstrating that mouse basigin can functionally replace fly basigin. To determine what regions of the basigin protein are required for each of these functions, we have created mutant basigin transgenes coding for proteins that are altered in conserved residues, introduced these into the fly genome, and tested them for their ability to rescue both photoreceptor cell structure defects and neuron-glia interaction defects of basigin. The results suggest that the highly conserved transmembrane domain and the extracellular domains are crucial for basigin function in the visual system while the short intracellular tail may not play a role in these functions.
Figures
Fig. 1
Sequence alignment of basigin related proteins. Identical residues are highlighted in black and similar residues are highlighted in yellow. Only the C-terminus of Drosophila basigin 298 is shown (last line). Drosophila basigin 298 (accession number CG31605-PG) can be found at NCBI or
. The thick black underline indicates the putative transmembrane sequence. Residues that were changed in altered basigin proteins are marked with an asterisk. When more than one residue was changed at a time this is indicated by a line above residues marked with asterisks.
Fig. 2
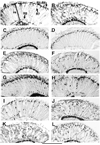
Testing basigin transgenes for their ability to rescue photoreceptor cell structural defects of _bsg_265. Frozen longitudinal head sections were labeled with anti-elav to visualize photoreceptor nuclear placement in the retina. re=retina. (A) Mosaic animal mutant in the eye for _bsg_265 but expressing wild-type basigin from an engineered transgene. Photoreceptor nuclei are placed as in wild-type animals (not shown). Labeled arrows show R1–R6, R7, and R8 nuclear placement. (B) Mosaic animals mutant in the eye for _bsg_265. Photoreceptor nuclei are found scattered throughout the retina. (C–L) Mosaic animal mutant in the eye for _bsg_265 but expressing the following basigin mutant proteins. (C) YE246SG. (D) PFL228LFTL. Photoreceptor nuclear placement is normal. (E) T128A. (F) RVK218LVT. (G) EG132GV. (H) WKK64LKT. (I) Y103S. (J) E235G. (K) W151G. (L) DRGEY188AVRES.
Fig. 3
Examination of protein stability from basigin mutant transgenes by immunoblot with anti-V5antibody. Our transgenic lines all make basigin protein with a V5 terminal tag. We expressed mutant proteins in the eye via an ey-Gal4 driver and performed an immunoblot with anti-V5 antibody. Arrows indicate the location of two MW markers. The proteins represented in order are (1) EG132GV. (2) T128A. (3) WKK64LKT. (4) EIE113GFG. (5) RVK218LVT. (6) wild-type basigin. (7) DRGEY188AVRES. (8) E235G. (9) W151G. (10) Truncated V5. Arrows show the positions of the nearest MW markers.
Fig. 4
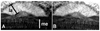
Expression of protein from Drosophila basigin transgenes in the visual system. Frozen longitudinal head sections were labeled with anti-basigin antibody. (A) Mosaic animal mutant in the eye for _bsg_265 but expressing basigin E235G from an engineered transgene. la= lamina, me=medulla. (B) Mosaic animal mutant in the eye for _bsg_265 but expressing basigin W151G from an engineered transgene. The expression pattern is the same as wild-type basigin (Curtin et al., 2007).
Fig. 5
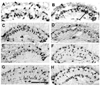
Testing basigin mutant transgenes for their ability to rescue neuron-glia interaction defects of bsg265 Frozen longitudinal head sections were labeled with anti-repo, an antibody specific to glia cell nuclei. The post-synaptic lamina is shown. la=lamina. Three distinct layers of glia can be ascertained, each marked with distinctive arrows in panels (A) and (B). The arrow with the long stem and small head points to the epithelial glia, the ones found altered when the eye is mutant for _bsg_265. (A) Mosaic animal mutant in the eye for _bsg_265 show scattered epithelia glia. (B) Mosaic animals mutant in the eye for _bsg_265 but expressing wild-type basigin in the eye from an engineered transgene. Glia cell placement is like wild-type animals (not shown). ( C) Mosaic animal mutant in the eye for _bsg_265 but expressing mutant basigin protein YE246SG from a transgene. Glia cell placement is normal. (D) Mosaic animal mutant in the eye for _bsg_265 but expressing mutant basigin protein P147L from a transgene. Glia cell placement is normal. (E) Mosaic animal mutant in the eye for _bsg_265 but expressing mutant basigin protein PFL228LFT from a transgene. Glia cell placement is like _bsg_265 . (F) Mosaic animal mutant in the eye for _bsg_265 but expressing mutant basigin protein EIE113GFG from a transgene. Glia cell placement is like _bsg_265 . (G) Mosaic animal mutant in the eye for _bsg_265 but expressing mutant basigin protein E235G from a transgene. Epithelia glia are located at the proximal margin of the lamina. (H) Mosaic animal mutant in the eye for _bsg_265 but expressing mutant basigin protein W151G from a transgene. Epithelia glia are located at the proximal margin of the lamina.
Similar articles
- Basigin/EMMPRIN/CD147 mediates neuron-glia interactions in the optic lamina of Drosophila.
Curtin KD, Wyman RJ, Meinertzhagen IA. Curtin KD, et al. Glia. 2007 Nov 15;55(15):1542-53. doi: 10.1002/glia.20568. Glia. 2007. PMID: 17729283 - Basigin (EMMPRIN/CD147) interacts with integrin to affect cellular architecture.
Curtin KD, Meinertzhagen IA, Wyman RJ. Curtin KD, et al. J Cell Sci. 2005 Jun 15;118(Pt 12):2649-60. doi: 10.1242/jcs.02408. Epub 2005 May 31. J Cell Sci. 2005. PMID: 15928045 - Basigin Associates with Integrin in Order to Regulate Perineurial Glia and Drosophila Nervous System Morphology.
Hunter AC, Petley-Ragan LM, Das M, Auld VJ. Hunter AC, et al. J Neurosci. 2020 Apr 22;40(17):3360-3373. doi: 10.1523/JNEUROSCI.1397-19.2020. Epub 2020 Apr 7. J Neurosci. 2020. PMID: 32265259 Free PMC article. - Basigin (CD147): a multifunctional transmembrane protein involved in reproduction, neural function, inflammation and tumor invasion.
Muramatsu T, Miyauchi T. Muramatsu T, et al. Histol Histopathol. 2003 Jul;18(3):981-7. doi: 10.14670/HH-18.981. Histol Histopathol. 2003. PMID: 12792908 Review. - Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners.
Muramatsu T. Muramatsu T. J Biochem. 2016 May;159(5):481-90. doi: 10.1093/jb/mvv127. Epub 2015 Dec 18. J Biochem. 2016. PMID: 26684586 Free PMC article. Review.
Cited by
- A cell surface biotinylation assay to reveal membrane-associated neuronal cues: Negr1 regulates dendritic arborization.
Pischedda F, Szczurkowska J, Cirnaru MD, Giesert F, Vezzoli E, Ueffing M, Sala C, Francolini M, Hauck SM, Cancedda L, Piccoli G. Pischedda F, et al. Mol Cell Proteomics. 2014 Mar;13(3):733-48. doi: 10.1074/mcp.M113.031716. Epub 2013 Dec 31. Mol Cell Proteomics. 2014. PMID: 24382801 Free PMC article. - Modulation of miR-210 alters phasing of circadian locomotor activity and impairs projections of PDF clock neurons in Drosophila melanogaster.
Cusumano P, Biscontin A, Sandrelli F, Mazzotta GM, Tregnago C, De Pittà C, Costa R. Cusumano P, et al. PLoS Genet. 2018 Jul 16;14(7):e1007500. doi: 10.1371/journal.pgen.1007500. eCollection 2018 Jul. PLoS Genet. 2018. PMID: 30011269 Free PMC article. - Basolateral sorting signals regulating tissue-specific polarity of heteromeric monocarboxylate transporters in epithelia.
Castorino JJ, Deborde S, Deora A, Schreiner R, Gallagher-Colombo SM, Rodriguez-Boulan E, Philp NJ. Castorino JJ, et al. Traffic. 2011 Apr;12(4):483-98. doi: 10.1111/j.1600-0854.2010.01155.x. Epub 2011 Feb 1. Traffic. 2011. PMID: 21199217 Free PMC article.
References
- Altruda F, Cervella P, Gaeta ML, et al. Cloning of cDNA for a novel mouse membrane glycoprotein (gp 42): shared identity to histocompatibility antigens, immunoglobulins and neural-cell adhesion molecules. Gene. 1989;85:445–452. - PubMed
- Anholt RR, Dilda CL, Chang S, Fanara JJ, et al. The genetic architecture of odor-guided behavior in Drosophila: epistasis and the transcriptome. Nature Genetics. 2003;35:180–184. - PubMed
- Aplin AE, Howe A, Alahari SK, et al. Signal transduction and signal modulation by cell adhesion receptors: the role of integrins, cadherins, immunoglobulin-cell adhesion molecules, and selectins. Pharmacol. Rev. 1998;50:197–264. - PubMed
- Berditchevski F, Chang S, Bodorova J, et al. Generation of monoclonal antibodies to integrin-associated proteins. Evidence that alpha3beta1 complexes with EMMPRIN/basigin/OX47/M6. J. Biol. Chem. 1997;272:29174–29180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous