Exploring molecular mechanisms of ligand recognition by opioid receptors with metadynamics - PubMed (original) (raw)
Exploring molecular mechanisms of ligand recognition by opioid receptors with metadynamics
Davide Provasi et al. Biochemistry. 2009.
Abstract
Opioid receptors are G protein-coupled receptors (GPCRs) of utmost significance in the development of potent analgesic drugs for the treatment of severe pain. An accurate evaluation at the molecular level of the ligand binding pathways into these receptors may play a key role in the design of new molecules with more desirable properties and reduced side effects. The recent characterization of high-resolution X-ray crystal structures of non-rhodopsin GPCRs for diffusible hormones and neurotransmitters presents an unprecedented opportunity to build improved homology models of opioid receptors, and to study in more detail their molecular mechanisms of ligand recognition. In this study, possible pathways for entry of the nonselective antagonist naloxone (NLX) from the water environment into the well-accepted alkaloid binding pocket of a delta opioid receptor (DOR) molecular model based on the beta2-adrenergic receptor crystal structure are explored using microsecond-scale well-tempered metadynamics simulations. Using as collective variables distances that account for the position of NLX and of the receptor extracellular loop 2 in relation to the DOR binding pocket, we were able to distinguish between the different states visited by the ligand (i.e., docked, undocked, and metastable bound intermediates) and to predict a free energy of binding close to experimental values after correcting for possible drawbacks of the sampling approach. The strategy employed herein holds promise for its application to the docking of diverse ligands to the opioid receptors as well as to other GPCRs.
Figures
Figure 1
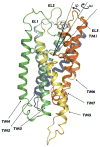
Side view of the initial 3D model of human DOR built according to the procedure described in Methods. TM1, TM2, TM3, TM4, TM5, TM6, and TM7 are colored in purple, blue, light blue, light green, green, yellow, orange and red, respectively. The 5-degree conical restraint that was applied to circumscribe NLX sampling is indicated by the black lines defining the angle between the COM of the binding pocket, the COM of the ligand, and the COM of residue L3007.35 (black dot in the figure).
Figure 2
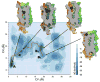
Free-energy surface of the NLX-DOR system reconstructed by well-tempered metadynamics as a function of the distance of the NLX COM (CV1) and of the distance of EL2 COM (CV2) from the binding pocket COM. Relevant states are labeled A (NLX bound into the well-accepted OR alkaloid binding pocket), B (NLX bound at the EL2/EL3 recognition cleft), and C (NLX at a metastable state in the helix bundle). Each contour represents a free-energy difference of 2 kcal/mol. The red solid line refers to the entry path obtained by NEB that was used to generate the entry path collective variables. Also represented are images of A, B1, B2, and C metastable states of DOR cut along their TM4 face and the position of NLX (black spheres) in the corresponding states.
Figure 3
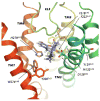
Representative conformation extracted from the basin A of the free-energy surface showing NLX bound to the DOR well-accepted alkaloid binding pocket. Parts of TM2 and TM4 have been removed for clarity.
Figure 4
Representative conformations extracted from basins B1, B2 and C of the free-energy surface. Top (as seen from the extracellular side) and side views of NLX bound (A, B) to the EL2/EL3 cleft on the DOR surface in the conformation extracted from the B1 basin, (C, D) to the EL2/EL3 cleft on the DOR surface in the conformation extracted from the B2 basin, and (E, F) within the helix bundle, in the conformation extracted from the C basin.
Figure 5
The free-energy surface reconstructed using well-tempered metadynamics as a function of the position along (CV3) and the distance from (CV4) the suggested NLX entry path. Each contour represents a free-energy difference of 2 kcal/mol. Relevant states are labeled according to Figure 2, and are: A (NLX bound to the DOR well-accepted alkaloid binding pocket, see Figure 3), B1 (NLX bound to the most external location on the EL2/EL3 cleft, see Figures 4A,B), B2 (NLX bound to a more stable position on the EL2/EL3 cleft, see Figures 4C,D) and C (NLX at a metastable state within the helix bundle).
Figure 6
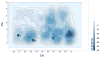
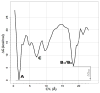
Integration of the free-energy profile of Figure 2 over the CV2 variable (distance of the EL2 C198-W209 region from the receptor alkaloid binding pocket) reported as a function of the CV1 variable (distance of NLX from the receptor alkaloid binding pocket) in the 0<CV1<20 region. The free-energy profile was shifted so that the reference state corresponds to the most stable one.
Similar articles
- Exploring the structure of opioid receptors with homology modeling based on single and multiple templates and subsequent docking: a comparative study.
Bera I, Laskar A, Ghoshal N. Bera I, et al. J Mol Model. 2011 May;17(5):1207-21. doi: 10.1007/s00894-010-0803-8. Epub 2010 Jul 27. J Mol Model. 2011. PMID: 20661609 - Mechanistic insights into the allosteric modulation of opioid receptors by sodium ions.
Shang Y, LeRouzic V, Schneider S, Bisignano P, Pasternak GW, Filizola M. Shang Y, et al. Biochemistry. 2014 Aug 12;53(31):5140-9. doi: 10.1021/bi5006915. Epub 2014 Jul 29. Biochemistry. 2014. PMID: 25073009 Free PMC article. - 3D modeling, ligand binding and activation studies of the cloned mouse delta, mu; and kappa opioid receptors.
Filizola M, Laakkonen L, Loew GH. Filizola M, et al. Protein Eng. 1999 Nov;12(11):927-42. doi: 10.1093/protein/12.11.927. Protein Eng. 1999. PMID: 10585498 - [Structure of mu and delta opioid receptors].
Granier S. Granier S. Med Sci (Paris). 2012 Oct;28(10):870-5. doi: 10.1051/medsci/20122810016. Epub 2012 Oct 12. Med Sci (Paris). 2012. PMID: 23067419 Review. French. - Efficiency of Homology Modeling Assisted Molecular Docking in G-protein Coupled Receptors.
Bhunia SS, Saxena AK. Bhunia SS, et al. Curr Top Med Chem. 2021;21(4):269-294. doi: 10.2174/1568026620666200908165250. Curr Top Med Chem. 2021. PMID: 32901584 Review.
Cited by
- Action of molecular switches in GPCRs--theoretical and experimental studies.
Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A, Filipek S. Trzaskowski B, et al. Curr Med Chem. 2012;19(8):1090-109. doi: 10.2174/092986712799320556. Curr Med Chem. 2012. PMID: 22300046 Free PMC article. Review. - A multiscale predictive digital twin for neurocardiac modulation.
Yang PC, Rose A, DeMarco KR, Dawson JRD, Han Y, Jeng MT, Harvey RD, Santana LF, Ripplinger CM, Vorobyov I, Lewis TJ, Clancy CE. Yang PC, et al. J Physiol. 2023 Sep;601(17):3789-3812. doi: 10.1113/JP284391. Epub 2023 Aug 1. J Physiol. 2023. PMID: 37528537 Free PMC article. - Energy Landscapes of Ligand Motion Inside the Tunnel-Like Cavity of Lipid Transfer Proteins: The Case of the Pru p 3 Allergen.
Cuevas-Zuviría B, Garrido-Arandia M, Díaz-Perales A, Pacios LF. Cuevas-Zuviría B, et al. Int J Mol Sci. 2019 Mar 21;20(6):1432. doi: 10.3390/ijms20061432. Int J Mol Sci. 2019. PMID: 30901853 Free PMC article. - Mechanistic Understanding from Molecular Dynamics in Pharmaceutical Research 2: Lipid Membrane in Drug Design.
Róg T, Girych M, Bunker A. Róg T, et al. Pharmaceuticals (Basel). 2021 Oct 19;14(10):1062. doi: 10.3390/ph14101062. Pharmaceuticals (Basel). 2021. PMID: 34681286 Free PMC article. Review. - Thermodynamics of camphor migration in cytochrome P450cam by atomistic simulations.
Rydzewski J, Nowak W. Rydzewski J, et al. Sci Rep. 2017 Aug 10;7(1):7736. doi: 10.1038/s41598-017-07993-0. Sci Rep. 2017. PMID: 28798338 Free PMC article.
References
- Befort K, Tabbara L, Bausch S, Chavkin C, Evans C, Kieffer B. The conserved aspartate residue in the third putative transmembrane domain of the delta-opioid receptor is not the anionic counterpart for cationic opiate binding but is a constituent of the receptor binding site. Mol Pharmacol. 1996;49:216–223. - PubMed
- Surratt CK, Johnson PS, Moriwaki A, Seidleck BK, Blaschak CJ, Wang JB, Uhl GR. -mu opiate receptor. Charged transmembrane domain amino acids are critical for agonist recognition and intrinsic activity. J Biol Chem. 1994;269:20548–20553. - PubMed
- Li JG, Chen C, Yin J, Rice K, Zhang Y, Matecka D, de Riel JK, DesJarlais RL, Liu-Chen LY. ASP147 in the third transmembrane helix of the rat mu opioid receptor forms ion-pairing with morphine and naltrexone. Life Sci. 1999;65:175–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DA020032-05/DA/NIDA NIH HHS/United States
- K02 DA026434-01/DA/NIDA NIH HHS/United States
- R01 DA020032/DA/NIDA NIH HHS/United States
- DA026434/DA/NIDA NIH HHS/United States
- K02 DA026434/DA/NIDA NIH HHS/United States
- DA020032/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources