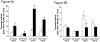Associations of a polymorphism in the ornithine decarboxylase gene with colorectal cancer survival - PubMed (original) (raw)
Associations of a polymorphism in the ornithine decarboxylase gene with colorectal cancer survival
Jason A Zell et al. Clin Cancer Res. 2009.
Abstract
Purpose: Activity of ornithine decarboxylase (ODC), the first enzyme in polyamine synthesis, is required for normal growth and is elevated in many cancers, including colorectal cancer. We examined associations of the +316 ODC1 single nucleotide polymorphism (SNP) with colorectal cancer-specific survival among colorectal cancer cases, and then investigated its functional significance in colon cancer cells.
Experimental design: The study included 400 incident stage I-III colorectal cancer cases from the population-based University of California Irvine Gene-Environment Study of Familial Colorectal Cancer (diagnosed from 1994 to 1996 with follow-up through March 2008). The primary outcome was colorectal cancer-specific survival dependent on ODC1 (rs2302615) genotype (GG versus GA/AA). In human colon cancer cell lines, ODC1 allele-specific binding of E-box transcription factors was determined via Western blotting and chromatin immunoprecipitation assays. ODC1 allele-specific promoter activity was determined using promoter constructs in combination with vectors expressing either the transcriptional activator c-MYC or the repressor MAD1.
Results: Genotype-specific survival differences were observed among colorectal cancer cases: compared with cases with the ODC1 GG genotype (hazards ratio, 1; reference) the adjusted colorectal cancer-specific survival hazards ratio was 2.02 (95% confidence interval, 1.17-3.50) for ODC1 GA/AA cases (P = 0.012). In colon cancer cells, the ODC1 SNP, flanked by two E-boxes, predicts ODC1 promoter activity. The E-box activator c-MYC and repressors MAD1 and MAD4 preferentially bind to ODC1 minor A-alleles, compared with major G-alleles, in cultured cells.
Conclusions: These results have implications for conditional regulation of polyamine homeostasis and suggest a model in which the ODC1 SNP may be protective for colon adenoma recurrence and detrimental for survival after colon cancer diagnosis.
Conflict of interest statement
Disclosure of Potential Conflict of Interest: Eugene W. Gerner has an ownership interest in Cancer Prevention Pharmaceuticals, LLC.
Figures
Fig. 1
Schema depicting the proposed differential effects of polyamine regulation by MAD1 and c-MYC on the ornithine decarboxylase-1 (ODC-1) +316 minor A-allele. Effects of the ODC inhibitor DFMO (difluoromethylornithine) are also shown.
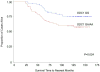
Fig. 2
Kaplan-Meier colorectal cancer (CRC) specific survival rate estimates for cases with stage III CRC, stratified by ornithine decarboxylase 1 (ODC1) +316 genotype. Includes cases from the University of California Irvine Gene-Environment Study of Familial CRC diagnosed during 1994-1996 with follow-up through March 2008; ODC1 GG (64 cases, 15 CRC-specific deaths), ODC1 GA/AA (62 cases, 25 CRC-specific deaths).
Fig. 3
A) Location of the ODC1 promoter SNP. The SNP under investigation in this study is 316 nucleotides 3′ of the ODC1 transcription start site (marked with an asterisk). This SNP resides between two consensus E-boxes as shown by the underlined sequences, and affects a PstI restriction site, marked with a box. B) RFLP analysis of ODC1 SNP. DNA was obtained from two cell types, and the region surrounding the ODC1 SNP site was sequenced. Colon-derived HT29 cells were found to be heterozygous GA, while HCT116 cells were found to be homozygous GG, at the ODC1 SNP locus. A 350 bp PCR product of this region was obtained from each cell type and subjected to digestion with PstI. Evidence of an A-allele was indicated by restriction products smaller than 350 bp.
Fig. 3
A) Location of the ODC1 promoter SNP. The SNP under investigation in this study is 316 nucleotides 3′ of the ODC1 transcription start site (marked with an asterisk). This SNP resides between two consensus E-boxes as shown by the underlined sequences, and affects a PstI restriction site, marked with a box. B) RFLP analysis of ODC1 SNP. DNA was obtained from two cell types, and the region surrounding the ODC1 SNP site was sequenced. Colon-derived HT29 cells were found to be heterozygous GA, while HCT116 cells were found to be homozygous GG, at the ODC1 SNP locus. A 350 bp PCR product of this region was obtained from each cell type and subjected to digestion with PstI. Evidence of an A-allele was indicated by restriction products smaller than 350 bp.
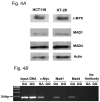
Fig. 4
A) E-box protein expression in colon-derived cells. Expression of proteins to be evaluated for binding to the +316 ODC1 SNP was assessed by Western blot analysis. Extracts of both HT29 and HCT116 cells were evaluated for c-MYC, MAD1 and MAD4; β-actin was used as a loading control. B) Documentation of allele-specific transcription factor binding by chromatin immunoprecipitation (CHIP) analysis. CHIP analysis was conducted as described in Material and Methods. HT29 cells were a source of ODC1 A-alleles, as these cells are heterozygous GA at this site. HCT116 cells were used as a source of ODC1 G-alleles.
Fig. 5
A) Effect of c-MYC expression on ODC1 allele-specific promoter activity in HT29 colon derived cells. Promoter activity was measured after transfection with ODC1 promoter reporter plasmids co-transfected with pcDNA 3.0 plasmid (□) or CMV-MYC expression vector (■). Promoter constructs differ by the presence of the first E-box element, located in −485 to −480 bp (“wt E-box1” for the wild-type sequence or “mut E-box1”, for a mutant sequence). The constructs differ also by the ODC1 +316 SNP (“+316 G” or “+316 A”). *, P ≤ 0.013 for each of the four comparisons relative to promoter activity with pcDNA 3.0 co-transfection. B) Effect of MAD1 expression on ODC1 allele-specific promoter activity in HT29 colon tumor derived cells. Promoter activity was measured after transfection with ODC1 promoter reporter plasmids co-transfected with pcDNA 3.1 plasmid (□) or with a pcDNA-_MAD_1 plasmid (■). Promoter constructs used were described in the legend for panel A of this figure. *, P = 0.027, statistical significance relative to promoter activity with pcDNA 3.1 co-transfection.
Similar articles
- Pronounced reduction in adenoma recurrence associated with aspirin use and a polymorphism in the ornithine decarboxylase gene.
Martinez ME, O'Brien TG, Fultz KE, Babbar N, Yerushalmi H, Qu N, Guo Y, Boorman D, Einspahr J, Alberts DS, Gerner EW. Martinez ME, et al. Proc Natl Acad Sci U S A. 2003 Jun 24;100(13):7859-64. doi: 10.1073/pnas.1332465100. Epub 2003 Jun 16. Proc Natl Acad Sci U S A. 2003. PMID: 12810952 Free PMC article. - Association between the ornithine decarboxylase G316A polymorphism and breast cancer survival.
Xu L, Long J, Wang P, Liu K, Mai L, Guo Y. Xu L, et al. Oncol Lett. 2015 Jul;10(1):485-491. doi: 10.3892/ol.2015.3201. Epub 2015 May 12. Oncol Lett. 2015. PMID: 26171056 Free PMC article. - Ornithine decarboxylase-1 polymorphism, chemoprevention with eflornithine and sulindac, and outcomes among colorectal adenoma patients.
Zell JA, McLaren CE, Chen WP, Thompson PA, Gerner EW, Meyskens FL. Zell JA, et al. J Natl Cancer Inst. 2010 Oct 6;102(19):1513-6. doi: 10.1093/jnci/djq325. Epub 2010 Aug 26. J Natl Cancer Inst. 2010. PMID: 20798393 Free PMC article. Clinical Trial. - Meat consumption, ornithine decarboxylase gene polymorphism, and outcomes after colorectal cancer diagnosis.
Zell JA, Lin BS, Ziogas A, Anton-Culver H. Zell JA, et al. J Carcinog. 2012;11:17. doi: 10.4103/1477-3163.104004. Epub 2012 Nov 28. J Carcinog. 2012. PMID: 23233821 Free PMC article. - Ornithine decarboxylase G316A genotype is prognostic for colorectal adenoma recurrence and predicts efficacy of aspirin chemoprevention.
Hubner RA, Muir KR, Liu JF, Logan RF, Grainge MJ, Houlston RS; Members of the UKCAP Consortium. Hubner RA, et al. Clin Cancer Res. 2008 Apr 15;14(8):2303-9. doi: 10.1158/1078-0432.CCR-07-4599. Clin Cancer Res. 2008. PMID: 18413818
Cited by
- DFMO: targeted risk reduction therapy for colorectal neoplasia.
Laukaitis CM, Gerner EW. Laukaitis CM, et al. Best Pract Res Clin Gastroenterol. 2011 Aug;25(4-5):495-506. doi: 10.1016/j.bpg.2011.09.007. Best Pract Res Clin Gastroenterol. 2011. PMID: 22122766 Free PMC article. Review. - Difluoromethylornithine (DFMO) and Neuroblastoma: A Review.
Tangella AV, Gajre AS, Chirumamilla PC, Rathhan PV. Tangella AV, et al. Cureus. 2023 Apr 17;15(4):e37680. doi: 10.7759/cureus.37680. eCollection 2023 Apr. Cureus. 2023. PMID: 37206500 Free PMC article. Review. - Variants downstream of the ornithine decarboxylase gene influence risk of colorectal adenoma and aspirin chemoprevention.
Barry EL, Mott LA, Sandler RS, Ahnen DJ, Baron JA. Barry EL, et al. Cancer Prev Res (Phila). 2011 Dec;4(12):2072-82. doi: 10.1158/1940-6207.CAPR-11-0300. Epub 2011 Sep 19. Cancer Prev Res (Phila). 2011. PMID: 21930798 Free PMC article. Clinical Trial. - A Phase I Trial of DFMO Targeting Polyamine Addiction in Patients with Relapsed/Refractory Neuroblastoma.
Saulnier Sholler GL, Gerner EW, Bergendahl G, MacArthur RB, VanderWerff A, Ashikaga T, Bond JP, Ferguson W, Roberts W, Wada RK, Eslin D, Kraveka JM, Kaplan J, Mitchell D, Parikh NS, Neville K, Sender L, Higgins T, Kawakita M, Hiramatsu K, Moriya SS, Bachmann AS. Saulnier Sholler GL, et al. PLoS One. 2015 May 27;10(5):e0127246. doi: 10.1371/journal.pone.0127246. eCollection 2015. PLoS One. 2015. PMID: 26018967 Free PMC article. Clinical Trial. - Lack of evidence for the association of ornithine decarboxylase (+316 G>A), spermidine/spermine acetyl transferase (-1415 T>C) gene polymorphisms with calcium oxalate stone disease.
Coker-Gürkan A, Arisan S, Arisan ED, Unsal NP. Coker-Gürkan A, et al. Biomed Rep. 2014 Jan;2(1):69-74. doi: 10.3892/br.2013.184. Epub 2013 Oct 21. Biomed Rep. 2014. PMID: 24649071 Free PMC article.
References
- Wallace HM. The physiological role of the polyamines. Eur J Clin Invest. 2000;30:1–3. - PubMed
- Gerner EW, Meyskens FL. Polyamines and cancer: Old molecules, new understanding. Nature Reviews Cancer. 2004;4:781–92. - PubMed
- Giardiello FM, Hamilton SR, Hylind LM, Yang VW, Tamez P, Casero RA. Ornithine decarboxylase and polyamines in familial adenomatous polyposis. Cancer Research. 1997;57:199–201. - PubMed
- Fultz KE, Gerner EW. APC-dependent regulation of ornithine decarboxylase in human colon tumor cells. MolCarcinog. 2002;34:10–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 CA095060-03S1/CA/NCI NIH HHS/United States
- P50 CA095060-05S3/CA/NCI NIH HHS/United States
- U01 CA078285-05S2/CA/NCI NIH HHS/United States
- CA72008/CA/NCI NIH HHS/United States
- P50 CA095060-070001/CA/NCI NIH HHS/United States
- U01 CA078285-05S1/CA/NCI NIH HHS/United States
- K23 CA133142/CA/NCI NIH HHS/United States
- P50 CA095060-06A19003/CA/NCI NIH HHS/United States
- P01 CA072008-08/CA/NCI NIH HHS/United States
- P01 CA072008-01A10001/CA/NCI NIH HHS/United States
- U01 CA078285-03/CA/NCI NIH HHS/United States
- P01 CA072008-05/CA/NCI NIH HHS/United States
- P01 CA072008-03S3/CA/NCI NIH HHS/United States
- U01 CA078285-05/CA/NCI NIH HHS/United States
- U24 CA078134-02S1/CA/NCI NIH HHS/United States
- P50 CA095060-05S2/CA/NCI NIH HHS/United States
- CA95060/CA/NCI NIH HHS/United States
- U01 CA078285-02/CA/NCI NIH HHS/United States
- N01-PC-35139/PC/NCI NIH HHS/United States
- P50 CA095060-02/CA/NCI NIH HHS/United States
- P50 CA095060-04/CA/NCI NIH HHS/United States
- P01 CA072008-020001/CA/NCI NIH HHS/United States
- U01 CA078285-04/CA/NCI NIH HHS/United States
- P50 CA095060-080001/CA/NCI NIH HHS/United States
- CA78285/CA/NCI NIH HHS/United States
- L30 CA130160/CA/NCI NIH HHS/United States
- P50 CA095060-08/CA/NCI NIH HHS/United States
- P01 CA072008-07/CA/NCI NIH HHS/United States
- P50 CA095060-03/CA/NCI NIH HHS/United States
- P01 CA072008-03S2/CA/NCI NIH HHS/United States
- U24 CA078134-02/CA/NCI NIH HHS/United States
- P01 CA072008-030001/CA/NCI NIH HHS/United States
- U01 CA078285-05S3/CA/NCI NIH HHS/United States
- P01 CA072008-03S10001/CA/NCI NIH HHS/United States
- P50 CA095060/CA/NCI NIH HHS/United States
- P01 CA072008-04A10001/CA/NCI NIH HHS/United States
- P50 CA095060-05/CA/NCI NIH HHS/United States
- U58 DP000807/DP/NCCDPHP CDC HHS/United States
- P01 CA072008-06S1/CA/NCI NIH HHS/United States
- P50 CA095060-07S1/CA/NCI NIH HHS/United States
- P50 CA095060-05S1/CA/NCI NIH HHS/United States
- P01 CA072008-04A1/CA/NCI NIH HHS/United States
- U01 CA078285-05S4/CA/NCI NIH HHS/United States
- N01PC54404/CA/NCI NIH HHS/United States
- L30 CA130160-01/CA/NCI NIH HHS/United States
- P01 CA072008-06/CA/NCI NIH HHS/United States
- P01 CA072008-03S1/CA/NCI NIH HHS/United States
- CA78134/CA/NCI NIH HHS/United States
- P50 CA095060-01/CA/NCI NIH HHS/United States
- K23 CA133142-01A1/CA/NCI NIH HHS/United States
- P01 CA072008-07S1/CA/NCI NIH HHS/United States
- P01 CA072008/CA/NCI NIH HHS/United States
- P01 CA072008-03S30001/CA/NCI NIH HHS/United States
- P50 CA095060-010001/CA/NCI NIH HHS/United States
- P01 CA072008-08S1/CA/NCI NIH HHS/United States
- P50 CA095060-04S1/CA/NCI NIH HHS/United States
- P01 CA072008-03S20001/CA/NCI NIH HHS/United States
- P01 CA072008-02S10001/CA/NCI NIH HHS/United States
- N01PC35136/CA/NCI NIH HHS/United States
- R01 CA063706-05/CA/NCI NIH HHS/United States
- U01 CA078285-05S5/CA/NCI NIH HHS/United States
- P01 CA072008-03/CA/NCI NIH HHS/United States
- N01PC35139/CA/NCI NIH HHS/United States
- N01-PC-54404/PC/NCI NIH HHS/United States
- P50 CA095060-08S1/CA/NCI NIH HHS/United States
- P01 CA072008-050001/CA/NCI NIH HHS/United States
- U01 CA078285-02S1/CA/NCI NIH HHS/United States
- P50 CA095060-06A1/CA/NCI NIH HHS/United States
- P50 CA095060-06A10001/CA/NCI NIH HHS/United States
- N01-PC-35136/PC/NCI NIH HHS/United States
- P50 CA095060-07/CA/NCI NIH HHS/United States
- U24 CA078134/CA/NCI NIH HHS/United States
- U01 CA078285/CA/NCI NIH HHS/United States
- 1U58DP00807-01/DP/NCCDPHP CDC HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials