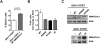The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart - PubMed (original) (raw)
The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart
Alberto Ciccia et al. Genes Dev. 2009.
Abstract
The integrity of genomic DNA is continuously challenged by the presence of DNA base lesions or DNA strand breaks. Here we report the identification of a new DNA damage response protein, SMARCAL1 (SWI/SNF-related, matrix associated, actin-dependent regulator of chromatin, subfamily a-like 1), which is a member of the SNF2 family and is mutated in Schimke immunoosseous dysplasia (SIOD). We demonstrate that SMARCAL1 directly interacts with Replication protein A (RPA) and is recruited to sites of DNA damage in an RPA-dependent manner. SMARCAL1-depleted cells display sensitivity to DNA-damaging agents that induce replication fork collapse, and exhibit slower fork recovery and delayed entry into mitosis following S-phase arrest. Furthermore, SIOD patient fibroblasts reconstituted with SMARCAL1 exhibit faster cell cycle progression after S-phase arrest. Thus, the symptoms of SIOD may be caused, at least in part, by defects in the cellular response to DNA replication stress.
Figures
Figure 1.
Interaction between SMARCAL1 and RPA in human cell lines. (A) Input cell lysates and HA immunoprecipitates from 293T-Rex cells expressing control HA vector, HA-RPA1, or HA-SMARCAL1 were immunoblotted with antibodies to HA, SMARCAL1, RPA1, and RPA2. Cells were treated with 10 Gy IR or 30 J/m2 UV or left untreated. (B) Protein complexes immunoprecipitated with IgG or anti-SMARCAL1 antibodies from 293T-Rex were analyzed for SMARCAL1 and RPA2 by Western blotting. (C) Protein complexes immunoprecipitated from 293T-Rex cells with IgG or anti-RPA2 antibodies were immunoblotted with antibodies to SMARCAL1, RPA1, and RPA2.
Figure 2.
In vitro interaction between SMARCAL1 and RPA. (A) Western blot of protein complexes pulled down with nickel beads from BL21 (DE3) bacteria expressing untagged RPA1, RPA2, or RPA3, with or without His-tagged SMARCAL1. (B) Sequence alignment of the putative RPA2-interacting motif of SMARCAL1 with the previously known RPA2-interacting motifs of TIPIN, UNG2, XPA, and RAD52. Similar residues are indicated within boxes. Sequence alignments were performed with ClustalW. The schematic representation of human SMARCAL1 and the RQK and ΔN mutants is shown below the sequence alignments. The amino acids mutated to alanine in the RQK mutant are indicated by asterisks. (C) Western blot of protein complexes pulled down from bacteria coexpressing RPA1, RPA2, and RPA3 with nickel beads in the presence of either wild-type, RQK, or ΔN His-tagged SMARCAL1. (D) HA immunoprecipitates from 293T-Rex cells expressing wild-type, RQK, or ΔN HA-SMARCAL1 were immunoblotted with antibodies to HA and RPA2.
Figure 3.
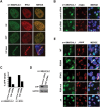
Colocalization of SMARCAL1 and RPA at DNA damage sites. (A) U2OS cells expressing GFP-SMARCAL1 were stained with anti-GFP (green) and anti-RPA2 (red) antibodies following treatment with 2 mM HU, 10 nM CPT, or microirradiation with UV laser. (B) GFP-SMARCAL1 U2OS cells transfected with control or CtIP siRNAs were stained with anti-GFP (green) and anti-γH2AX (red) antibodies following UV laser microirradiation. Merged images have DAPI staining (blue). (C) Percentage of control or CtIP siRNA-treated U2OS cells displaying GFP-SMARCAL1 or GFP-RPA1 stripes over the total number of cells with γH2AX stripes following UV laser microirradiation. The data represent the average of two independent experiments. (D) Cell lysates from cells described in C were immunoblotted for CtIP and GAPDH. (E) U2OS cells expressing wild-type and mutant GFP-SMARCAL1 constructs were microirradiated with a UV laser and stained as in B.
Figure 4.
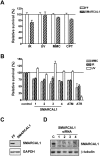
DNA damage sensitivity of SMARCAL1-depleted cells. (A) Cell competition assay showing survival of U2OS cells infected with SMARCAL1 shRNA or FF shRNA retroviruses, relative to dsRed-expressing U2OS cells, following treatment with DNA-damaging agents. (IR) 5 Gy IR; (UV) 7 J/m2 UV; (MMC) 150 nM MMC; (CPT) 10 nM CPT. (B) Cell competition assay using siRNAs targeting SMARCAL1, ATM, or ATR, or a nontargeting control siRNA. Treatments are the same as described in A. (C) Western blot showing depletion of SMARCAL1 in cells infected with retrovirus expressing SMARCAL1 shRNA. (D) Western blot showing knockdown of SMARCAL1 in U2OS cells transfected with four individual SMARCAL1 siRNAs.
Figure 5.
Effects of SMARCAL1 depletion on cell cycle progression. (A) Analysis of FF control and SMARCAL1-depleted U2OS cells 10 h after a BrdU pulse alone (not treated, NT), or a BrdU pulse followed immediately by IR (2 Gy) or addition of CPT (5 nM). Nocodazole was added after the BrdU pulse to capture mitotic cells, which are measured by staining for P-H3. Shown are the percentages of BrdU+ cells that are also P-H3+. Values are means and standard deviations from at least two experiments. _P_-values were calculated using a one-tailed _t_-test. (B) Analysis of FF control and SMARCAL1-depleted U2OS cells 10 h after a BrdU pulse followed by 2 Gy IR or addition of 5 nM CPT in the presence of 5 mM caffeine. The analysis was performed as described in A. The numbers indicate the percentage of BrdU+ P-H3+ cells. (C) Analysis of mitotic cells (P-H3+) 10 h after release from the thymidine block. Propidium iodide was used to measure DNA content. Numbers indicate the percentage of P-H3+ cells.
Figure 6.
Effects of SMARCAL1 depletion on replication fork restart. (A) Restart of DNA replication in FF control or SMARCAL1 shRNA-depleted U2OS was measured by incorporation of BrdU following release from the thymidine block. BrdU (10 μM) was added 30 min prior to fixation. 7-AAD was used to measure DNA content. The percentage of BrdU+ cells is indicated. Results are representative of three independent experiments. (B) Schematics of the pulse-labeling experiment for DNA fiber analysis. (C) DNA fibers were stained with antibodies recognizing IdU (green) or CldU (red). Anti-ssDNA antibody was used as a control. (D) Analysis of the percentage of stalled or collapsed replication forks (green tracts only) in FF control or SMARCAL1 shRNA-depleted U2OS cells with or without aphidicolin treatment. The percentage of new origins fired during CldU treatment (red tracts only) is also shown.
Figure 7.
Analysis of cell cycle progression in SIOD patient fibroblasts reconstituted with SMARCAL1. (A) DNA replication restart in hTERT-immortalized SIOD patient SD31 fibroblasts and SD31 fibroblasts reconstituted with untagged SMARCAL1 was measured by incorporation of BrdU 3 h after release from a thymidine block. Results are means and standard deviations of two independent experiments. _P_-value was calculated using a two-tailed _t_-test. (B) Analysis of cell cycle progression in hTERT-immortalized SD31 fibroblasts and SD31 fibroblasts reconstituted with untagged wild-type or mutant SMARCAL1 constructs following a thymidine block. The graph shows the fold change in P-H3+ cells relative to nonreconstituted hTERT SD31 fibroblasts 15 h after release from a thymidine block into nocodazole. Values are means and standard deviations from at least two experiments. (*) P = 0.003. The _P_-value was calculated using a two-tailed _t_-test. (C) Western blot showing the expression of wild-type and mutant SMARCAL1 protein in the reconstituted hTERT-immortalized SD31 fibroblasts.
Comment in
- HARPing on about the DNA damage response during replication.
Driscoll R, Cimprich KA. Driscoll R, et al. Genes Dev. 2009 Oct 15;23(20):2359-65. doi: 10.1101/gad.1860609. Genes Dev. 2009. PMID: 19833762 Free PMC article.
Similar articles
- The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks.
Bansbach CE, Bétous R, Lovejoy CA, Glick GG, Cortez D. Bansbach CE, et al. Genes Dev. 2009 Oct 15;23(20):2405-14. doi: 10.1101/gad.1839909. Epub 2009 Sep 30. Genes Dev. 2009. PMID: 19793861 Free PMC article. - SMARCAL1 and replication stress: an explanation for SIOD?
Bansbach CE, Boerkoel CF, Cortez D. Bansbach CE, et al. Nucleus. 2010 May-Jun;1(3):245-8. doi: 10.4161/nucl.1.3.11739. Epub 2010 Feb 16. Nucleus. 2010. PMID: 21327070 Free PMC article. - [SMARCAL1, roles and mechanisms in genome stability maintenance].
Wen YL, Lü KN, Xu XK, Zhang X, Ding L, Pan XF. Wen YL, et al. Yi Chuan. 2019 Dec 20;41(12):1084-1098. doi: 10.16288/j.yczz.19-158. Yi Chuan. 2019. PMID: 31857280 Review. Chinese. - A novel splice site mutation in SMARCAL1 results in aberrant exon definition in a child with Schimke immunoosseous dysplasia.
Carroll C, Hunley TE, Guo Y, Cortez D. Carroll C, et al. Am J Med Genet A. 2015 Oct;167A(10):2260-4. doi: 10.1002/ajmg.a.37146. Epub 2015 May 5. Am J Med Genet A. 2015. PMID: 25943327 Free PMC article. - The role of SMARCAL1 in replication fork stability and telomere maintenance.
Lugli N, Sotiriou SK, Halazonetis TD. Lugli N, et al. DNA Repair (Amst). 2017 Aug;56:129-134. doi: 10.1016/j.dnarep.2017.06.015. Epub 2017 Jun 10. DNA Repair (Amst). 2017. PMID: 28623093 Review.
Cited by
- Mechanisms of direct replication restart at stressed replisomes.
Conti BA, Smogorzewska A. Conti BA, et al. DNA Repair (Amst). 2020 Nov;95:102947. doi: 10.1016/j.dnarep.2020.102947. Epub 2020 Aug 16. DNA Repair (Amst). 2020. PMID: 32853827 Free PMC article. Review. No abstract available. - Chromatin and the genome integrity network.
Papamichos-Chronakis M, Peterson CL. Papamichos-Chronakis M, et al. Nat Rev Genet. 2013 Jan;14(1):62-75. doi: 10.1038/nrg3345. Nat Rev Genet. 2013. PMID: 23247436 Free PMC article. Review. - HARPing on about the DNA damage response during replication.
Driscoll R, Cimprich KA. Driscoll R, et al. Genes Dev. 2009 Oct 15;23(20):2359-65. doi: 10.1101/gad.1860609. Genes Dev. 2009. PMID: 19833762 Free PMC article. - Smarcal1 and Zranb3 Protect Replication Forks from Myc-Induced DNA Replication Stress.
Puccetti MV, Adams CM, Kushinsky S, Eischen CM. Puccetti MV, et al. Cancer Res. 2019 Apr 1;79(7):1612-1623. doi: 10.1158/0008-5472.CAN-18-2705. Epub 2019 Jan 4. Cancer Res. 2019. PMID: 30610086 Free PMC article. - On the Interaction Between SMARCAL1 and BRG1.
Bisht D, Patne K, Rakesh R, Muthuswami R. Bisht D, et al. Front Cell Dev Biol. 2022 Jun 16;10:870815. doi: 10.3389/fcell.2022.870815. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35784471 Free PMC article.
References
- Boerkoel CF, Takashima H, John J, Yan J, Stankiewicz P, Rosenbarker L, Andre JL, Bogdanovic R, Burguet A, Cockfield S, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet. 2002;30:215–220. - PubMed
- Coleman MA, Eisen JA, Mohrenweiser HW. Cloning and characterization of HARP/SMARCAL1: A prokaryotic HepA-related SNF2 helicase protein from human and mouse. Genomics. 2000;65:274–282. - PubMed
- Collins N, Poot RA, Kukimoto I, Garcia-Jimenez C, Dellaire G, Varga-Weisz PD. An ACF1–ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat Genet. 2002;32:627–632. - PubMed
- Deguchi K, Clewing JM, Elizondo LI, Hirano R, Huang C, Choi K, Sloan EA, Lucke T, Marwedel KM, Powell RD, Jr, et al. Neurologic phenotype of Schimke immuno-osseous dysplasia and neurodevelopmental expression of SMARCAL1. J Neuropathol Exp Neurol. 2008;67:565–577. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG011085/AG/NIA NIH HHS/United States
- R01 CA107342/CA/NCI NIH HHS/United States
- R01 GM094972/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials