The duration of Fgf8 isthmic organizer expression is key to patterning different tectal-isthmo-cerebellum structures - PubMed (original) (raw)
. 2009 Nov;136(21):3617-26.
doi: 10.1242/dev.041210. Epub 2009 Sep 30.
Affiliations
- PMID: 19793884
- PMCID: PMC2761110
- DOI: 10.1242/dev.041210
The duration of Fgf8 isthmic organizer expression is key to patterning different tectal-isthmo-cerebellum structures
Tatsuya Sato et al. Development. 2009 Nov.
Abstract
The isthmic organizer and its key effector molecule, fibroblast growth factor 8 (Fgf8), have been cornerstones in studies of how organizing centers differentially pattern tissues. Studies have implicated different levels of Fgf8 signaling from the mid/hindbrain boundary (isthmus) as being responsible for induction of different structures within the tectal-isthmo-cerebellum region. However, the role of Fgf8 signaling for different durations in patterning tissues has not been studied. To address this, we conditionally ablated Fgf8 in the isthmus and uncovered that prolonged expression of Fgf8 is required for the structures found progressively closer to the isthmus to form. We found that cell death cannot be the main factor accounting for the loss of brain structures near the isthmus, and instead demonstrate that tissue transformation underlies the observed phenotypes. We suggest that the remaining Fgf8 and Fgf17 signaling in our temporal Fgf8 conditional mutants is sufficient to ensure survival of most midbrain/hindbrain cells near the isthmus. One crucial role for sustained Fgf8 function is in repressing Otx2 in the hindbrain, thereby allowing the isthmus and cerebellum to form. A second requirement for sustained Fgf8 signaling is to induce formation of a posterior tectum. Finally, Fgf8 is also required to maintain the borders of expression of a number of key genes involved in tectal-isthmo-cerebellum development. Thus, the duration as well as the strength of Fgf8 signaling is key to patterning of the mid/hindbrain region. By extrapolation, the length of Fgf8 expression could be crucial to Fgf8 function in other embryonic organizers.
Figures
Fig. 1.
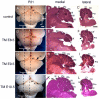
Adult brain morphology shows that Fgf8 is required for different lengths of time to form tectal-isthmo-cerebellum substructures. Dorsal view of wholemount adult brains (A,D,G,J), and Hematoxylin and Eosin-stained paramedial (B,E,H,K) and lateral (C,F,I,L) sections of Fgf8 temporal CKO embryos treated with TM at the time points indicated. The control is an_Fgf8flox/+; En2CreER/+_ embryo treated with TM at E8.5. Arrow in F points to bulge in simplex of Fgf8-E8.5 CKOs. Arrow in G points to loss of IC in Fgf8-E9.5 CKOs. Arrow in K points to decreased size of the IC in Fgf8-E10.5 CKOs. Dashed lines in wholemounts indicate the level of the indicated sections. A, anterior; cer, cerebellum; CI, crus I; CII, crus II; H, hemisphere; IC, inferior colliculus; I-X, vermis lobules, hemisphere lobules; S, simplex; SC, superior colliculus; P, copula pyramidis; Pm, paramedianus; Pv, paravermis; V, vermis. Scale bars: 1 mm.
Fig. 2.
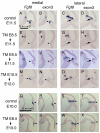
Recombination is efficient by 36 hours following TM in Fgf8 temporal CKOs. (A-X) RNA in situ hybridization with the two_Fgf8_ probes indicated on adjacent medial or lateral sections of_Fgf8_ temporal CKO embryos treated with TM at the time-points indicated. The ages of the embryos analyzed are indicated under the arrows at the left. The control embryo is Fgf8flox/+; En2CreER/+ treated with TM at E8.5. The transcripts detected with the Fgf8 and exon 3 probes were greatly diminished in the dorsomedial isthmus (arrowheads). (K,L,O,P) Although the Fgf8 mutant transcript was detected in the dorsolateral isthmus of Fgf8-E9.5 CKO and Fgf8-E10.5 CKOs, the normal transcript was greatly diminished (arrows). (E-H, asterisks) The normal Fgf8 transcript was only partially reduced ventrally. (N,P,V,X, arrowheads) The normal Fgf8 transcript was greatly diminished by 36 hours after TM administration in the dorsal isthmus. Scale bars: in A, 500 μm for A-P; in Q, 500 μm for Q-X.
Fig. 3.
Loss of Fgf18 expression and reduction of Fgf17 and_Sprty1_ expression soon after Fgf8 is ablated. (A-X) RNA in situ hybridization with the probes indicated of adjacent medial or lateral sections in each column of Fgf8 temporal CKO embryos treated with TM at the time-points indicated. The ages of the embryos analyzed are indicated to the right of the arrows at the top. The control is an Fgf8flox/+; En2CreER/+ embryo treated with TM at E8.5. Arrowheads indicate the expressions of Spry1 (A,C,E,G),Fgf17 (I,K,M,O) and Fgf18 (Q,S,U,W) in the dorsal-medial part of caudal midbrain-isthmus-r1 region. Scale bar: 500 μm.
Fig. 4.
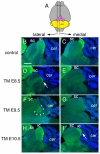
Neurogranin staining of adult brain sections shows that the inferior colliculus requires sustained Fgf8 signaling. (A) Schematic of dorsal view of an adult brain to show the level of the lateral and medial sections shown below. (B-I) Neurogranin staining of the inferior colliculus of adult Fgf8 temporal CKO embryos treated with TM at the time-points indicated. The control is an Fgf8flox/+; En2CreER/+ embryo treated with TM at E8.5. The arrow in D indicates the faint and abnormally positioned neurogranin staining in the_Fgf8-E8.5_ CKO. Arrowheads in F indicate the abnormal neurogranin staining in the lateral IC of a Fgf8-E9.5 CKO mutant. Arrow in I indicates the smaller medial IC in Fgf8-E10.5 CKO mice. Abbreviations are as in Fig. 1. Scale bar: 500 μm.
Fig. 5.
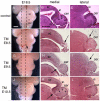
E18.5 brain morphology shows that Fgf8 is required for different lengths of time to form tectal-isthmo-cerebellum substructures. Dorsal view of wholemount adult brains (A,D,G,J), and Hematoxylin and Eosin-stained paramedial (medial; B,E,H,K) and lateral (C,F,I,L) sections of Fgf8 temporal CKO embryos treated with TM at the time-points indicated. The control is an Fgf8flox/+; En2CreER/+ embryo. Dashed lines in wholemounts indicate the level of the indicated sections. Arrows in D and E indicate loss of the vermis, in addition to the inferior colliculus in_Fgf8-E8.5_ CKO mutants, and in F the shallow isthmus flexure. The arrow in H indicates the lack of isthmus and inferior colliculus in_Fgf8-E9.5_ CKO mutants, and in I the shallow isthmus flexure. The arrow in K indicates that the isthmus is missing in Fgf8-E10.5 CKO mutants, in addition to the cerebellum fissures being shallower than normal (B,F, asterisks). Abbreviations are as in Fig. 1. Scale bars: in A, 1 mm for left column; in B, 500 μm for right two columns.
Fig. 6.
Apoptosis is not greatly increased near the dorsal isthmus when_Fgf8_ is ablated by ∼E10. Dorsal (A,B,E,F) and lateral (C,D,G,H) views of E10 control (A-D;Fgf8flox/+; En2CreER/+, TM at E8.5) and_Fgf8-E8.5_ CKO embryos (E-H) stained with Lysotracker. Arrows in E and F indicate a slight increase in apoptotic cells in the dorsal midline of the r1 in mutants. Arrowheads in G and H indicate an increase in apoptosis in the diencephalon/mesencephalic border region of mutants. di, diencephalons; mes, mesencephalon; r1, rhombomere1.
Fig. 7.
Otx2 and Wnt1 are expanded posteriorly and_Gbx2_ is diminished soon after Fgf8 is ablated. (A-P′) RNA in situ hybridization with the probes indicated of adjacent medial sections in each column of Fgf8 temporal CKO embryos treated with TM at the time-points indicated. The ages of the embryos analyzed are indicated under the arrows at the top. The control is an_Fgf8flox/+; En2CreER/+_ embryo treated with TM at E8.5. Rectangles indicate area shown at higher magnification to the right. Green asterisks indicate caudal limit of the Otx2 expression. Red asterisks indicate rostral limit of the Fgf8 expression. Arrows in A-H indicate that Otx2 and Wnt1 are expanded posteriorly in the dorsal isthmus-r1 of mutants. (I-P) Dorsal Fgf8 and Gbx2 expression are diminished in mutants (arrowheads). Scale bar: in A, 300 μm for A-P; in A′, 300 μm for A′-P′.
Fig. 8.
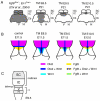
Schematic showing the progressive loss of structures and more severe changes in gene expression near the isthmus as Fgf8 is ablated earlier. (A) Back view of adult brain showing cerebellum [medial vermis (V) and lateral hemispheres (H)] and tectum (IC and SC). Shading indicates the regions of the tectum and cerebellum that are lost when_Fgf8_ is conditionally ablated from the isthmus at the times indicated or at E8.5 in Fgf8flox-; En1Cre embryos (Chi et al., 2004). The isthmus is lost in all the mutants (not shown). (B) Changes in gene expression seen at the times indicated in Fgf8 temporal CKO embryos. Fgf8 indicates the mutant transcript. (C) Dorsal view of E9.5 neural tube showing the primordial for the SC, IC, isthmus, vermis (V) and hemispheres (H).
Similar articles
- Lmx1b is essential for Fgf8 and Wnt1 expression in the isthmic organizer during tectum and cerebellum development in mice.
Guo C, Qiu HY, Huang Y, Chen H, Yang RQ, Chen SD, Johnson RL, Chen ZF, Ding YQ. Guo C, et al. Development. 2007 Jan;134(2):317-25. doi: 10.1242/dev.02745. Epub 2006 Dec 13. Development. 2007. PMID: 17166916 - Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling.
Blaess S, Stephen D, Joyner AL. Blaess S, et al. Development. 2008 Jun;135(12):2093-103. doi: 10.1242/dev.015990. Epub 2008 May 14. Development. 2008. PMID: 18480159 Free PMC article. - FGF8 induces formation of an ectopic isthmic organizer and isthmocerebellar development via a repressive effect on Otx2 expression.
Martinez S, Crossley PH, Cobos I, Rubenstein JL, Martin GR. Martinez S, et al. Development. 1999 Mar;126(6):1189-200. doi: 10.1242/dev.126.6.1189. Development. 1999. PMID: 10021338 - Isthmus organizer for midbrain and hindbrain development.
Nakamura H, Katahira T, Matsunaga E, Sato T. Nakamura H, et al. Brain Res Brain Res Rev. 2005 Sep;49(2):120-6. doi: 10.1016/j.brainresrev.2004.10.005. Epub 2005 Jan 21. Brain Res Brain Res Rev. 2005. PMID: 16111543 Review. - Fgf8 signaling for development of the midbrain and hindbrain.
Harada H, Sato T, Nakamura H. Harada H, et al. Dev Growth Differ. 2016 Jun;58(5):437-45. doi: 10.1111/dgd.12293. Epub 2016 Jun 7. Dev Growth Differ. 2016. PMID: 27273073 Review.
Cited by
- Revisiting the development of cerebellar inhibitory interneurons in the light of single-cell genetic analyses.
Schilling K. Schilling K. Histochem Cell Biol. 2024 Jan;161(1):5-27. doi: 10.1007/s00418-023-02251-z. Epub 2023 Nov 8. Histochem Cell Biol. 2024. PMID: 37940705 Free PMC article. Review. - The embryonic patterning gene Dbx1 governs the survival of the auditory midbrain via Tcf7l2-Ap2δ transcriptional cascade.
Tran HN, Nguyen QH, Jeong JE, Loi DL, Nam YH, Kang TH, Yoon J, Baek K, Jeong Y. Tran HN, et al. Cell Death Differ. 2023 Jun;30(6):1563-1574. doi: 10.1038/s41418-023-01165-6. Epub 2023 Apr 20. Cell Death Differ. 2023. PMID: 37081114 Free PMC article. - Transcription factors regulating the specification of brainstem respiratory neurons.
Xia Y, Cui K, Alonso A, Lowenstein ED, Hernandez-Miranda LR. Xia Y, et al. Front Mol Neurosci. 2022 Nov 29;15:1072475. doi: 10.3389/fnmol.2022.1072475. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36523603 Free PMC article. Review. - FGF8-FGFR1 signaling regulates human GnRH neuron differentiation in a time- and dose-dependent manner.
Yellapragada V, Eskici N, Wang Y, Madhusudan S, Vaaralahti K, Tuuri T, Raivio T. Yellapragada V, et al. Dis Model Mech. 2022 Aug 1;15(8):dmm049436. doi: 10.1242/dmm.049436. Epub 2022 Aug 16. Dis Model Mech. 2022. PMID: 35833364 Free PMC article. - An Update on the Molecular Mechanism of the Vertebrate Isthmic Organizer Development in the Context of the Neuromeric Model.
Hidalgo-Sánchez M, Andreu-Cervera A, Villa-Carballar S, Echevarria D. Hidalgo-Sánchez M, et al. Front Neuroanat. 2022 Mar 24;16:826976. doi: 10.3389/fnana.2022.826976. eCollection 2022. Front Neuroanat. 2022. PMID: 35401126 Free PMC article. Review.
References
- Acampora, D., Mazan, S., Lallemand, Y., Avantaggiato, V., Maury, M., Simeone, A. and Brûlet, P. (1995). Forebrain and midbrain regions are deleted in Otx2-/- mutants due to a defective anterior neuroectoderm specification during gastrulation. Development 121, 3279-3290. - PubMed
- Ang, S. L., Conlon, R. A., Jin, O. and Rossant, J. (1994). Positive and negative signals from mesoderm regulate the expression of mouse Otx2 in ectoderm explants. Development 120, 2979-2989. - PubMed
- Ang, S. L., Jin, O., Rhinn, M., Daigle, N., Stevenson, L. and Rossant, J. (1996). A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development 122, 243-252. - PubMed
- Blaess, S., Corrales, J. D. and Joyner, A. L. (2006). Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development 133, 1799-1809. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HD050767-02/HD/NICHD NIH HHS/United States
- R01 HD050767-01/HD/NICHD NIH HHS/United States
- R01 HD050767-05/HD/NICHD NIH HHS/United States
- R01 HD050767/HD/NICHD NIH HHS/United States
- R01 HD050767-04/HD/NICHD NIH HHS/United States
- R01 HD050767-03/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous