Metagenomic study of the oral microbiota by Illumina high-throughput sequencing - PubMed (original) (raw)
Metagenomic study of the oral microbiota by Illumina high-throughput sequencing
Vladimir Lazarevic et al. J Microbiol Methods. 2009 Dec.
Abstract
To date, metagenomic studies have relied on the utilization and analysis of reads obtained using 454 pyrosequencing to replace conventional Sanger sequencing. After extensively scanning the 16S ribosomal RNA (rRNA) gene, we identified the V5 hypervariable region as a short region providing reliable identification of bacterial sequences available in public databases such as the Human Oral Microbiome Database. We amplified samples from the oral cavity of three healthy individuals using primers covering an approximately 82-base segment of the V5 loop, and sequenced using the Illumina technology in a single orientation. We identified 135 genera or higher taxonomic ranks from the resulting 1,373,824 sequences. While the abundances of the most common phyla (Firmicutes, Proteobacteria, Actinobacteria, Fusobacteria and TM7) are largely comparable to previous studies, Bacteroidetes were less present. Potential sources for this difference include classification bias in this region of the 16S rRNA gene, human sample variation, sample preparation and primer bias. Using an Illumina sequencing approach, we achieved a much greater depth of coverage than previous oral microbiota studies, allowing us to identify several taxa not yet discovered in these types of samples, and to assess that at least 30,000 additional reads would be required to identify only one additional phylotype. The evolution of high-throughput sequencing technologies, and their subsequent improvements in read length enable the utilization of different platforms for studying communities of complex flora. Access to large amounts of data is already leading to a better representation of sample diversity at a reasonable cost.
Figures
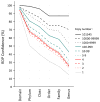
Fig. 1
Average confidence level for the six taxonomic levels as a function of sequence counts.
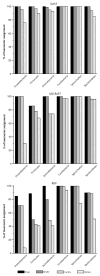
Fig. 2
Proportions of taxonomic assignments under the phylum level. Reads assigned to each of the four taxonomic levels for each major phylum are represented by bars. Their height represent the percentage of reads that can be placed at a given level of taxonomy using GAST, the MG-RAST server and the RDP Classifier.
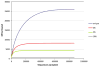
Fig. 3
Rarefaction analysis of the oral metagenome. The curves include only sequences which occur 3 or more times. The number of OTUs with different cutoff values was plotted as a function of the number of sequences sampled. OTUs with ≥97%, ≥95% and ≥90% pairwise sequence identity are arbitrarily assumed to form the same species, genus and family, respectively.
Similar articles
- The human oral microbiome.
Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. Dewhirst FE, et al. J Bacteriol. 2010 Oct;192(19):5002-17. doi: 10.1128/JB.00542-10. Epub 2010 Jul 23. J Bacteriol. 2010. PMID: 20656903 Free PMC article. - Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison.
Ahn J, Yang L, Paster BJ, Ganly I, Morris L, Pei Z, Hayes RB. Ahn J, et al. PLoS One. 2011;6(7):e22788. doi: 10.1371/journal.pone.0022788. Epub 2011 Jul 29. PLoS One. 2011. PMID: 21829515 Free PMC article. - Metagenomic analysis of the canine oral cavity as revealed by high-throughput pyrosequencing of the 16S rRNA gene.
Sturgeon A, Stull JW, Costa MC, Weese JS. Sturgeon A, et al. Vet Microbiol. 2013 Mar 23;162(2-4):891-898. doi: 10.1016/j.vetmic.2012.11.018. Epub 2012 Nov 20. Vet Microbiol. 2013. PMID: 23228621 - Insights into the human oral microbiome.
Verma D, Garg PK, Dubey AK. Verma D, et al. Arch Microbiol. 2018 May;200(4):525-540. doi: 10.1007/s00203-018-1505-3. Epub 2018 Mar 23. Arch Microbiol. 2018. PMID: 29572583 Review. - Profiling of Oral Bacterial Communities.
Wade WG, Prosdocimi EM. Wade WG, et al. J Dent Res. 2020 Jun;99(6):621-629. doi: 10.1177/0022034520914594. Epub 2020 Apr 14. J Dent Res. 2020. PMID: 32286907 Free PMC article. Review.
Cited by
- Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences.
Soergel DA, Dey N, Knight R, Brenner SE. Soergel DA, et al. ISME J. 2012 Jul;6(7):1440-4. doi: 10.1038/ismej.2011.208. Epub 2012 Jan 12. ISME J. 2012. PMID: 22237546 Free PMC article. - Culture and molecular-based profiles show shifts in bacterial communities of the upper respiratory tract that occur with age.
Stearns JC, Davidson CJ, McKeon S, Whelan FJ, Fontes ME, Schryvers AB, Bowdish DM, Kellner JD, Surette MG. Stearns JC, et al. ISME J. 2015 May;9(5):1246-59. doi: 10.1038/ismej.2014.250. Epub 2015 Jan 9. ISME J. 2015. PMID: 25575312 Free PMC article. - Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease.
Wang J, Qi J, Zhao H, He S, Zhang Y, Wei S, Zhao F. Wang J, et al. Sci Rep. 2013;3:1843. doi: 10.1038/srep01843. Sci Rep. 2013. PMID: 23673380 Free PMC article. - Diversity of the gut, vaginal and oral microbiome among pregnant women in South Africa with and without pre-eclampsia.
Geldenhuys J, Redelinghuys MJ, Lombaard HA, Ehlers MM, Cowan D, Kock MM. Geldenhuys J, et al. Front Glob Womens Health. 2022 Sep 16;3:810673. doi: 10.3389/fgwh.2022.810673. eCollection 2022. Front Glob Womens Health. 2022. PMID: 36188424 Free PMC article. - Defining the "core microbiome" of the microbial communities in the tonsils of healthy pigs.
Lowe BA, Marsh TL, Isaacs-Cosgrove N, Kirkwood RN, Kiupel M, Mulks MH. Lowe BA, et al. BMC Microbiol. 2012 Feb 7;12:20. doi: 10.1186/1471-2180-12-20. BMC Microbiol. 2012. PMID: 22313693 Free PMC article.
References
- Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA, Quan PL, Briese T, Hornig M, Geiser DM, Martinson V, vanEngelsdorp D, Kalkstein AL, Drysdale A, Hui J, Zhai J, Cui L, Hutchison SK, Simons JF, Egholm M, Pettis JS, Lipkin WI. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources