Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells - PubMed (original) (raw)
Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells
Daniela Meilinger et al. EMBO Rep. 2009 Nov.
Abstract
Recent studies have indicated that nuclear protein of 95 kDa (Np95) is essential for maintaining genomic methylation by recruiting DNA methyltransferase (Dnmt) 1 to hemi-methylated sites. Here, we show that Np95 interacts more strongly with regulatory domains of the de novo methyltransferases Dnmt3a and Dnmt3b. To investigate possible functions, we developed an epigenetic silencing assay using fluorescent reporters in embryonic stem cells (ESCs). Interestingly, silencing of the cytomegalovirus promoter in ESCs preceded DNA methylation and was strictly dependent on the presence of either Np95, histone H3 methyltransferase G9a or Dnmt3a and Dnmt3b. Our results indicate a regulatory role for Np95, Dnmt3a and Dnmt3b in mediating epigenetic silencing through histone modification followed by DNA methylation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
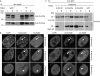
Np95 interacts with de novo methyltransferases Dnmt3a and Dnmt3b. (A) Co-immunoprecipitation of Dnmt3a and Dnmt3b with Np95 in wt and _np95_−/− E14 ESCs. The Dnmt3a2 isoform is shown in the lower panel. (B) F2H shows recruitment of Cherry–Np95 (prey) at the lac operator array (indicated by arrowheads) when GFP fusions of full-length Dnmt3a and Dnmt3b1 (G-Dnmt3a/b fl) or their amino-terminal regions (G-Dnmt3a/b N) are used as bait and not with their isolated C-terminal catalytic domains (G-Dnmt3a/b C). Scale bars, 5 μm. (C) Co-immunoprecipitation of Np95-His with GFP-tagged Dnmt1, Dnmt3a and Dnmt3b1 (G-Dnmt) transiently co-expressed in HEK293T cells. Co-expression of GFP was used as the control. In the upper row, immunoprecipitations carried out in the presence of 150 mM NaCl throughout the procedure are shown, whereas in the lower row, immunoprecipitation and wash buffers were carried out using 300 and 500 mM NaCl, respectively. Two per cent of input and supernatant relative to bound fractions were loaded in (A) and (C). B, bound; Dnmt, DNA methyltransferase; ESCs, embryonic stem cells; F2H, fluorescent two-hybrid assay; GFP, green fluorescent protein; HEK293T, human embryonic kidney 293T; I, input; Np95, nuclear protein of 95 kDa; S, supernatant; wt, wild type.
Figure 2
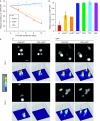
Promoter silencing activity in wild-type and mutant ESCs. ESCs with the indicated phenotypes were transiently co-transfected with CMV promoter-driven mRFP and CAG promoter-driven GFP reporter constructs. Between 90 and 150 images per sample were acquired either (A) every second day after transfection from a single experiment or (B) only at days 2 and 7–10 after transfection from 3–5 independent experiments. Relative levels of red over green fluorescence are shown with values for day 2 (first day of imaging) set to 1 (A). Wild-type J1 and E14 cells gave similar results (data not shown). _suv39_−/− stands for Suv39h1/2 double null ESCs. (C) Representative images of wt and _np95_−/− E14 ESCs co-transfected as in (A) and (B) (upper panels) and respective heat map intensity plots (lower panels). Scale bar, 15 μm. CAG, CMV early enhancer/chicken β actin promoter; CMV, cytomegalovirus promoter; DKO, double knockout; Dnmt, DNA methyltransferase; ESCs, embryonic stem cells; GFP, green fluorescent protein; mRFP, monomeric red fluorescent protein; Np95, nuclear protein of 95 kDa; TKO, triple knockout; wt, wild type.
Figure 3
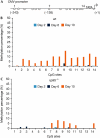
Methylation of the CMV promoter 2, 6 and 10 days after transfection. Wild-type and _np95_−/− ESCs were transfected as in Fig 2 and GFP-positive cells were sequentially sorted at the indicated days after transfection. Total DNA was isolated from sorted cells and bisulphite-treated. A proximal part of the CMV promoter was amplified and subjected to pyrosequencing. (A) Schematic drawing of the 14 proximal CpG sites analysed (shown as open circles). The numbers above correspond to the CpG sites shown in (B) and (C) and numbers in brackets refer to the position of CpG sites with respect to the transcription start site. (B,C) Methylation percentage at individual CpG sites for (B) wt and (C) _np95_−/− ESCs as measured by pyrosequencing. CMV, cytomegalovirus; ESCs, embryonic stem cells; GFP, green fluorescent protein; wt, wild type.
Similar articles
- The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA.
Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. Sharif J, et al. Nature. 2007 Dec 6;450(7171):908-12. doi: 10.1038/nature06397. Epub 2007 Nov 11. Nature. 2007. PMID: 17994007 - Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog.
Li JY, Pu MT, Hirasawa R, Li BZ, Huang YN, Zeng R, Jing NH, Chen T, Li E, Sasaki H, Xu GL. Li JY, et al. Mol Cell Biol. 2007 Dec;27(24):8748-59. doi: 10.1128/MCB.01380-07. Epub 2007 Oct 15. Mol Cell Biol. 2007. PMID: 17938196 Free PMC article. - The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome.
Gujar H, Weisenberger DJ, Liang G. Gujar H, et al. Genes (Basel). 2019 Feb 23;10(2):172. doi: 10.3390/genes10020172. Genes (Basel). 2019. PMID: 30813436 Free PMC article. Review. - Using human disease mutations to understand de novo DNA methyltransferase function.
Rolls W, Wilson MD, Sproul D. Rolls W, et al. Biochem Soc Trans. 2024 Oct 30;52(5):2059-2075. doi: 10.1042/BST20231017. Biochem Soc Trans. 2024. PMID: 39446312 Free PMC article. Review.
Cited by
- Prevotella copri exhausts intrinsic indole-3-pyruvic acid in the host to promote breast cancer progression: inactivation of AMPK via UHRF1-mediated negative regulation.
Su J, Lin X, Li D, Yang C, Lv S, Chen X, Yang X, Pan B, Xu R, Ren L, Zhang Y, Xie Y, Chen Q, Xia C. Su J, et al. Gut Microbes. 2024 Jan-Dec;16(1):2347757. doi: 10.1080/19490976.2024.2347757. Epub 2024 May 21. Gut Microbes. 2024. PMID: 38773738 Free PMC article. - Targeting the chromatin structural changes of antitumor immunity.
Li NN, Lun DX, Gong N, Meng G, Du XY, Wang H, Bao X, Li XY, Song JW, Hu K, Li L, Li SY, Liu W, Zhu W, Zhang Y, Li J, Yao T, Mou L, Han X, Hao F, Hu Y, Liu L, Zhu H, Wu Y, Liu B. Li NN, et al. J Pharm Anal. 2024 Apr;14(4):100905. doi: 10.1016/j.jpha.2023.11.012. Epub 2023 Nov 29. J Pharm Anal. 2024. PMID: 38665224 Free PMC article. Review. - Non-canonical functions of UHRF1 maintain DNA methylation homeostasis in cancer cells.
Yamaguchi K, Chen X, Rodgers B, Miura F, Bashtrykov P, Bonhomme F, Salinas-Luypaert C, Haxholli D, Gutekunst N, Aygenli BÖ, Ferry L, Kirsh O, Laisné M, Scelfo A, Ugur E, Arimondo PB, Leonhardt H, Kanemaki MT, Bartke T, Fachinetti D, Jeltsch A, Ito T, Defossez PA. Yamaguchi K, et al. Nat Commun. 2024 Apr 5;15(1):2960. doi: 10.1038/s41467-024-47314-4. Nat Commun. 2024. PMID: 38580649 Free PMC article. - A simultaneous knockout knockin genome editing strategy in HSPCs potently inhibits CCR5- and CXCR4-tropic HIV-1 infection.
Dudek AM, Feist WN, Sasu EJ, Luna SE, Ben-Efraim K, Bak RO, Cepika AM, Porteus MH. Dudek AM, et al. Cell Stem Cell. 2024 Apr 4;31(4):499-518.e6. doi: 10.1016/j.stem.2024.03.002. Cell Stem Cell. 2024. PMID: 38579682 - Epigenetics Mechanisms of Honeybees: Secrets of Royal Jelly.
Alhosin M. Alhosin M. Epigenet Insights. 2023 Nov 29;16:25168657231213717. doi: 10.1177/25168657231213717. eCollection 2023. Epigenet Insights. 2023. PMID: 38033464 Free PMC article. Review.
References
- Achour M et al. (2008) The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene 27: 2187–2197 - PubMed
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M (2008) Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455: 818–821 - PubMed
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S (2008) Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455: 822–825 - PubMed
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 - PubMed
- Bostick M, Kim JK, Esteve P-O, Clark A, Pradhan S, Jacobsen SE (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases