Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline - PubMed (original) (raw)
. 2009 Oct 23;36(2):231-44.
doi: 10.1016/j.molcel.2009.09.020. Epub 2009 Oct 1.
Masaki Shirayama, Darryl Conte Jr, Jessica Vasale, Pedro J Batista, Julie M Claycomb, James J Moresco, Elaine M Youngman, Jennifer Keys, Matthew J Stoltz, Chun-Chieh G Chen, Daniel A Chaves, Shenghua Duan, Kristin D Kasschau, Noah Fahlgren, John R Yates 3rd, Shohei Mitani, James C Carrington, Craig C Mello
Affiliations
- PMID: 19800275
- PMCID: PMC2776052
- DOI: 10.1016/j.molcel.2009.09.020
Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline
Weifeng Gu et al. Mol Cell. 2009.
Abstract
Endogenous small RNAs (endo-siRNAs) interact with Argonaute (AGO) proteins to mediate sequence-specific regulation of diverse biological processes. Here, we combine deep-sequencing and genetic approaches to explore the biogenesis and function of endo-siRNAs in C. elegans. We describe conditional alleles of the Dicer-related helicase, drh-3, that abrogate both RNA interference and the biogenesis of endo-siRNAs, called 22G-RNAs. DRH-3 is a core component of RNA-dependent RNA polymerase (RdRP) complexes essential for several distinct 22G-RNA systems. We show that, in the germline, one system is dependent on worm-specific AGOs, including WAGO-1, which localizes to germline nuage structures called P granules. WAGO-1 silences certain genes, transposons, pseudogenes, and cryptic loci. Finally, we demonstrate that components of the nonsense-mediated decay pathway function in at least one WAGO-mediated surveillance pathway. These findings broaden our understanding of the biogenesis and diversity of 22G-RNAs and suggest additional regulatory functions for small RNAs.
Figures
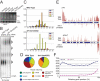
Figure 1. Hypomorphic alleles of drh-3 are RNAi deficient and temperature sensitive
A) Schematic of the drh-3 gene structure. Top panel: conserved DExH and HELICc domains and drh-3 lesions; bottom panel: four missense alleles (indicated) map to the HELICc domain. B) RNAi deficient phenotypes of drh-3 mutants. Fraction of viable embryos produced by animals exposed to pos-1 RNAi food, or viability of animals reared on let-2 RNAi food at 20°C. C and D) Brood size and Him phenotypes of drh-3 mutants at 20°C (dark bars) and 25°C (light bars). Mean brood size (top panel) of ≥10 hermaphrodites was determined by counting the number of embryos produced. Frequency of males among viable offspring was determined (bottom panel).
Figure 2. DRH-3 is required for the biogenesis of 22G-RNAs
A) Ethidium bromide staining of small RNAs. 22nt position is denoted by bracket. Note persistence of ~21nt band in all samples. 5.8S rRNA was used as loading control. B) Thin-layer chromatography analysis of first nucleotide of gel-purified 22 and 21nt RNA, as indicated, from wild type and drh-3 mutant samples. Bars (right), position of each nucleotide; “+”, treated with Terminator exonuclease; “−”, untreated. C) Length and first nucleotide distribution of genome matching reads from wild type and drh-3 mutant small RNA libraries. Sense structural RNAs were excluded in the analysis. D) Distribution of reads that match indicated genome annotations sequenced in wild type and drh-3 mutant small RNA libraries. E) Examples of small RNA distribution within rrf-1 and ama-1. Density of antisense reads indicated by vertical bars above (wild-type) or below (drh-3) gene structure. F) Genome-wide analysis of small RNA distribution within genes. Total antisense reads (y-axis) plotted according to the relative position (%) within all genes (x-axis; from 5′ to 3′).
Figure 3. Distinct requirements and genetic redundancy in 22G-RNA pathways
A) Northern blots of 22G-RNA expression in somatic gene Y47H10A.5, and germline targets F37D6.3 and Tc1 transposon in RNAi mutants. Loading controls: SL1 precursor and mir-66. B) Northern blots of germline 22G-RNAs in RdRP mutants. DA1316 is a congenic wild type control for the ego-1 and rrf-1 ego-1 mutants. Loading controls: SL1 precursor and mir-66. C) Coimmunoprecipitation analyses of DRH-3 from wild type and drh-3 lysates. Total lysate (left panels) and DRH-3 IPs (right panels) were analyzed by Western blot for candidate interacting proteins (indicated at right). The drh-3(0): the deletion allele (tm1217) control. Some DRH-3 protein detected in the drh-3(0) sample likely represents persistence of maternal product.
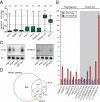
Figure 4. WAGO-1 and highly redundant WAGOs required for germline 22G-RNA biogenesis
A) Phylogenetic representation of WAGO proteins (see Supplemental Methods). WAGOs deleted in MAGO12 indicated in red. B), C) Northern blots of F37D6.3 22G-RNAs in multiple WAGO mutants. Loading control: SL1 precursor (B) and tRNA staining (C). D), E) Fluorescence microscopy of GFP::WAGO-1 and RFP::PGL-1, which colocalize to Pgranules in the germline.
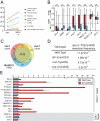
Figure 5. Deep-sequence analyses identified at least two distinct 22G-RNA pathways in the germline
A) Graph depicting change in reads matching indicated genome annotations between input and WAGO-1 IP samples. B) Change of 22G-RNAs derived from genes (red) or transposons (blue) in each mutant. Relative enrichment calculated as ratio of mutant / (mutant + wild type) for ‘n’ genes or transposons. WAGO-1 IP enrichment calculated as WAGO-1IP / (WAGO-1IP + IP input). C) Venn comparison of genes depleted of 22G-RNAs (≥ 2-fold) in indicated mutants (loci below the lower dashed line in (B)). D) Frequency of reversion of dpy-5::Tc5 as indicated (Ketting et al., 1999). E) Quantitative RT-PCR analysis of 22G-RNA target expression in drh-3 mutant (red) relative to wild-type (blue). WAGO and CSR-1 (gray) targets are indicated.
Figure 6. WAGO-associated 22G-RNAs define a surveillance system
A) Enrichment or depletion of 22G-RNAs derived from ‘n’ annotated pseudogenes. B) Quantitative RT-PCR analysis of pseudogenes and cryptic loci targeted by WAGO pathway in drh-3 mutant (red) relative to wild-type (blue). C) Northern blots of 22G-RNAs in smg mutants grown at 20°C. Similar results obtained with mutants grown at 25°C (not shown). Loading control: SL1 precursor. D) Venn comparison of genes depleted of 22G-RNAs (≥ 2-fold) in mutants.
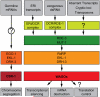
Figure 7
Model of germline 22G-RNA pathways required for genome surveillance in C. elegans.
Similar articles
- Caenorhabditis elegans RIG-I Homolog Mediates Antiviral RNA Interference Downstream of Dicer-Dependent Biogenesis of Viral Small Interfering RNAs.
Coffman SR, Lu J, Guo X, Zhong J, Jiang H, Broitman-Maduro G, Li WX, Lu R, Maduro M, Ding SW. Coffman SR, et al. mBio. 2017 Mar 21;8(2):e00264-17. doi: 10.1128/mBio.00264-17. mBio. 2017. PMID: 28325765 Free PMC article. - Widespread roles for piRNAs and WAGO-class siRNAs in shaping the germline transcriptome of Caenorhabditis elegans.
Reed KJ, Svendsen JM, Brown KC, Montgomery BE, Marks TN, Vijayasarathy T, Parker DM, Nishimura EO, Updike DL, Montgomery TA. Reed KJ, et al. Nucleic Acids Res. 2020 Feb 28;48(4):1811-1827. doi: 10.1093/nar/gkz1178. Nucleic Acids Res. 2020. PMID: 31872227 Free PMC article. - Protease-mediated processing of Argonaute proteins controls small RNA association.
Gudipati RK, Braun K, Gypas F, Hess D, Schreier J, Carl SH, Ketting RF, Großhans H. Gudipati RK, et al. Mol Cell. 2021 Jun 3;81(11):2388-2402.e8. doi: 10.1016/j.molcel.2021.03.029. Epub 2021 Apr 13. Mol Cell. 2021. PMID: 33852894 - Biology and Mechanisms of Short RNAs in Caenorhabditis elegans.
Grishok A. Grishok A. Adv Genet. 2013;83:1-69. doi: 10.1016/B978-0-12-407675-4.00001-8. Adv Genet. 2013. PMID: 23890211 Review. - Nuage condensates: accelerators or circuit breakers for sRNA silencing pathways?
Ouyang JPT, Seydoux G. Ouyang JPT, et al. RNA. 2022 Jan;28(1):58-66. doi: 10.1261/rna.079003.121. Epub 2021 Nov 12. RNA. 2022. PMID: 34772788 Free PMC article. Review.
Cited by
- Complex coding of endogenous siRNA, transcriptional silencing and H3K9 methylation on native targets of germline nuclear RNAi in C. elegans.
Ni JZ, Chen E, Gu SG. Ni JZ, et al. BMC Genomics. 2014 Dec 22;15(1):1157. doi: 10.1186/1471-2164-15-1157. BMC Genomics. 2014. PMID: 25534009 Free PMC article. - Functional diversification of Argonautes in nematodes: an expanding universe.
Buck AH, Blaxter M. Buck AH, et al. Biochem Soc Trans. 2013 Aug;41(4):881-6. doi: 10.1042/BST20130086. Biochem Soc Trans. 2013. PMID: 23863149 Free PMC article. Review. - Argonaute protein CSR-1 restricts localization of holocentromere protein HCP-3, the C. elegans CENP-A homolog.
Wong CYY, Tsui HN, Wang Y, Yuen KWY. Wong CYY, et al. J Cell Sci. 2024 Sep 15;137(18):jcs261895. doi: 10.1242/jcs.261895. Epub 2024 Sep 18. J Cell Sci. 2024. PMID: 39037215 Free PMC article. - Long noncoding RNAs in C. elegans.
Nam JW, Bartel DP. Nam JW, et al. Genome Res. 2012 Dec;22(12):2529-40. doi: 10.1101/gr.140475.112. Epub 2012 Jun 15. Genome Res. 2012. PMID: 22707570 Free PMC article. - Germ Granules Govern Small RNA Inheritance.
Lev I, Toker IA, Mor Y, Nitzan A, Weintraub G, Antonova O, Bhonkar O, Ben Shushan I, Seroussi U, Claycomb JM, Anava S, Gingold H, Zaidel-Bar R, Rechavi O. Lev I, et al. Curr Biol. 2019 Sep 9;29(17):2880-2891.e4. doi: 10.1016/j.cub.2019.07.054. Epub 2019 Aug 1. Curr Biol. 2019. PMID: 31378614 Free PMC article.
References
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. - PubMed
- Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS. 2007;581:2845–2853. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM063348/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM63348/GM/NIGMS NIH HHS/United States
- R01 DK074798/DK/NIDDK NIH HHS/United States
- R01 GM058800/GM/NIGMS NIH HHS/United States
- R01GM58800/GM/NIGMS NIH HHS/United States
- P41 RR011823/RR/NCRR NIH HHS/United States
- R37 GM058800/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials