Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport - PubMed (original) (raw)
Review
. 2009 Dec;8(12):1814-27.
doi: 10.1128/EC.00225-09. Epub 2009 Oct 2.
Affiliations
- PMID: 19801417
- PMCID: PMC2794212
- DOI: 10.1128/EC.00225-09
Review
Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport
Laura J Terry et al. Eukaryot Cell. 2009 Dec.
Abstract
The nuclear envelope is a physical barrier between the nucleus and cytoplasm and, as such, separates the mechanisms of transcription from translation. This compartmentalization of eukaryotic cells allows spatial regulation of gene expression; however, it also necessitates a mechanism for transport between the nucleus and cytoplasm. Macromolecular trafficking of protein and RNA occurs exclusively through nuclear pore complexes (NPCs), specialized channels spanning the nuclear envelope. A novel family of NPC proteins, the FG-nucleoporins (FG-Nups), coordinates and potentially regulates NPC translocation. The extensive repeats of phenylalanine-glycine (FG) in each FG-Nup directly bind to shuttling transport receptors moving through the NPC. In addition, FG-Nups are essential components of the nuclear permeability barrier. In this review, we discuss the structural features, cellular functions, and evolutionary conservation of the FG-Nups.
Figures
FIG. 1.
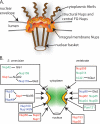
FG-Nups are distributed throughout the NPC. (A) Schematic representation of the eightfold radial symmetry of the NPC, showing key aspects of the NPC architecture. (B) Nup subcomplexes and relative NPC substructural localization. Each box represents a biochemically or functional documented subcomplex (from studies summarized by Alber et al. [3]). S. cerevisiae FG- Nups are depicted on the left side; vertebrates are depicted on the right. The FG-Nups (colored text) are found in discrete subcomplexes and substructural locations. This includes Nups containing predominantly FG (green text), GLFG (blue text), and FXFG (red text) repeats. Select structural, non-FG-Nups are shown in black text.
FIG. 2.
Key structural and sequence features of FG-domains in S. cerevisiae. (A) The full primary amino acid sequence of S. cerevisiae Nup49 is aligned with each FG repeat in a line break to align on the left. The FG repeats (green), FXFG (red), and GLFG (blue) are further highlighted. (B) Schematic diagrams for the 11 FG-Nups in S. cerevisiae showing the distribution and type of FG repeats. Single repeats are represented by an oval. FG repeat, green; FXFG repeat, red; GLFG repeat, blue. The diagrams are adapted from Strawn et al. (157) with permission from the publisher. (C) Structural analysis of an FXFG-importin β complex gives a surface view of the FXFG peptide (red) interaction pocket. Hydrophobic residues of importin β are highlighted in yellow. (Reprinted from reference with permission from the publisher.)
FIG. 3.
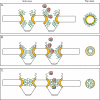
Models for the mechanism of NPC selectivity and transport. Based on the different features of the respective models, the distribution and physical features of the FG domains are distinct. This is represented in structural models of both a side view (perpendicular to the NE) and top view (cross-section through center of NPC; e.g., from cytoplasm onto plane of NE). NE, black; FG domains, green; structural NPC elements, yellow; importing karyopherin transport receptor, pink; NLS-bearing cargo, blue. (A) Brownian virtual gating model (138). The center of the NPC is a narrow channel, from which FG domains extend to form an entropic barrier to transport. Transport receptors bind these FG domains, overcoming the entropic barrier. By collecting on the NPC periphery, transport complexes increase the probability that they will spontaneously move across the barrier. (B) Reduction of Dimensionality model (125, 126). FG repeats form a continuous surface along the inner face of the NPC, and transport complexes pivot along this surface. The spacer sequence between FG repeats loop outward, forming a physical barrier to diffusion of large molecules; transport complexes might transiently displace these as they move along the FG surface. (C) Selective phase-partitioning model (133, 134). Hydrophobic interactions between FG repeats form a physical meshwork with gel-like properties. Transport receptors bind and transiently dissolve the meshwork in order to translocate through the NPC.
Similar articles
- Deciphering networks of protein interactions at the nuclear pore complex.
Allen NP, Patel SS, Huang L, Chalkley RJ, Burlingame A, Lutzmann M, Hurt EC, Rexach M. Allen NP, et al. Mol Cell Proteomics. 2002 Dec;1(12):930-46. doi: 10.1074/mcp.t200012-mcp200. Mol Cell Proteomics. 2002. PMID: 12543930 - 'Natively unfolded' nucleoporins in nucleocytoplasmic transport: clustered or evenly distributed?
Yang W. Yang W. Nucleus. 2011 Jan-Feb;2(1):10-6. doi: 10.4161/nucl.2.1.13818. Nucleus. 2011. PMID: 21647294 Free PMC article. - The mechanism of nucleocytoplasmic transport through the nuclear pore complex.
Tetenbaum-Novatt J, Rout MP. Tetenbaum-Novatt J, et al. Cold Spring Harb Symp Quant Biol. 2010;75:567-84. doi: 10.1101/sqb.2010.75.033. Epub 2011 Mar 29. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21447814 - The selective permeability barrier in the nuclear pore complex.
Li C, Goryaynov A, Yang W. Li C, et al. Nucleus. 2016 Sep 2;7(5):430-446. doi: 10.1080/19491034.2016.1238997. Epub 2016 Sep 27. Nucleus. 2016. PMID: 27673359 Free PMC article. Review. - The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport.
Wälde S, Kehlenbach RH. Wälde S, et al. Trends Cell Biol. 2010 Aug;20(8):461-9. doi: 10.1016/j.tcb.2010.05.001. Epub 2010 Jun 4. Trends Cell Biol. 2010. PMID: 20627572 Review.
Cited by
- Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane.
Ungricht R, Klann M, Horvath P, Kutay U. Ungricht R, et al. J Cell Biol. 2015 Jun 8;209(5):687-703. doi: 10.1083/jcb.201409127. J Cell Biol. 2015. PMID: 26056139 Free PMC article. - Disclosing the Interactome of Leukemogenic NUP98-HOXA9 and SET-NUP214 Fusion Proteins Using a Proteomic Approach.
Mendes A, Jühlen R, Bousbata S, Fahrenkrog B. Mendes A, et al. Cells. 2020 Jul 10;9(7):1666. doi: 10.3390/cells9071666. Cells. 2020. PMID: 32664447 Free PMC article. - Molecular interactions of FG nucleoporin repeats at high resolution.
Ibáñez de Opakua A, Geraets JA, Frieg B, Dienemann C, Savastano A, Rankovic M, Cima-Omori MS, Schröder GF, Zweckstetter M. Ibáñez de Opakua A, et al. Nat Chem. 2022 Nov;14(11):1278-1285. doi: 10.1038/s41557-022-01035-7. Epub 2022 Sep 22. Nat Chem. 2022. PMID: 36138110 Free PMC article. - The Palade symposium: celebrating cell biology at its best.
Schmid SL, Farquhar MG. Schmid SL, et al. Mol Biol Cell. 2010 Jul 15;21(14):2367-70. doi: 10.1091/mbc.e10-03-0179. Epub 2010 May 26. Mol Biol Cell. 2010. PMID: 20505070 Free PMC article. - Physical motif clustering within intrinsically disordered nucleoporin sequences reveals universal functional features.
Ando D, Colvin M, Rexach M, Gopinathan A. Ando D, et al. PLoS One. 2013 Sep 16;8(9):e73831. doi: 10.1371/journal.pone.0073831. eCollection 2013. PLoS One. 2013. PMID: 24066078 Free PMC article.
References
- Aitchison, J. D., G. Blobel, and M. P. Rout. 1996. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274:624-627. - PubMed
- Alber, F., S. Dokudovskaya, L. M. Veenhoff, W. Zhang, J. Kipper, D. Devos, A. Suprapto, O. Karni-Schmidt, R. Williams, B. T. Chait, A. Sali, and M. P. Rout. 2007. The molecular architecture of the nuclear pore complex. Nature 450:695-701. - PubMed
- Allen, N. P., L. Huang, A. Burlingame, and M. Rexach. 2001. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276:29268-29274. - PubMed
- Allen, N. P., S. S. Patel, L. Huang, R. J. Chalkley, A. Burlingame, M. Lutzmann, E. C. Hurt, and M. Rexach. 2002. Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell. Proteomics 1:930-946. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases