KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer - PubMed (original) (raw)
. 2009 Nov;11(11):1297-304.
doi: 10.1038/ncb1974. Epub 2009 Oct 4.
Affiliations
- PMID: 19801974
- PMCID: PMC2784164
- DOI: 10.1038/ncb1974
KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer
Kiranmai Gumireddy et al. Nat Cell Biol. 2009 Nov.
Abstract
Metastasis is a complex multistep process, which requires the concerted action of many genes and is the primary cause of cancer death. Both pathways that regulate metastasis enhancement and those that regulate its suppression contribute to the tumour dissemination process. To identify new metastasis suppressors, we set up a forward genetic screen in a mouse model. We transduced a genome-wide RNA interference (RNAi) library into the non-metastatic 168FARN breast cancer cell line and orthotopically transplanted the cells into mouse mammary fat pads. We then selected cells that could metastasize to the lung and identified an RNAi for the KLF17 gene. Conversely, we demonstrate that ectopic expression of KLF17 in a highly metastatic 4T1 breast cancer cell line inhibits the ability of cells to metastasize from the mammary fat pad to the lung. We also show that suppression of KLF17 expression promotes breast cancer cell invasion and epithelial-mesenchymal transition (EMT), and that KLF17 protein functions by directly binding to the promoter region of Id1 (which encodes a key metastasis regulator in breast cancer) to inhibit its transcription. Finally, we demonstrate that KLF17 expression is significantly downregulated in primary human breast cancer samples and that the combined expression pattern of KLF17 and Id1 can serve as a potential biomarker for lymph node metastasis in breast cancer.
Figures
Figure 1
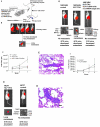
Identification of KLF17 as a metastasis-suppressing gene. (A) The scheme for the forward genetic screen in a mouse model. (B) Tumor cell growth in the primary site following injection of 168FARN-Luc cells containing a genome-wide RNAi library in mammary fat pad. One out of five mice developed lung metastasis. (C) Tumor growth in the primary sites following the transplantation of 168FARN cells expressing a control non-target shRNA; 168FARN expressing KLF17 shRNA; and 168FARN cells expressing both KLF17 shRNA and KLF17 cDNA in mammary fat pads. The growth rate is similar in all three groups before week 7. Tumor growth in 168FARN cells expressing KLF17 shRNA is higher than the other two groups at week 7 (p=0.001, t-test, n=10). (D) Transplantation of 168FARN cells stably expressing KLF17 shRNA in mammary fat pad leads to lung metastasis. Transplantation of 168FARN expressing a non-target shRNA does not lead to metastasis. No metastasis developed following the transplantation of 168FARN cells stably expressing both KLF17 shRNA and KLF17 cDNA which lacks shRNA binding site. (E) Histology analysis of metastasis in the lung following the transplantation of 168FARN cells expressing KLF17 shRNA. Arrows indicate micrometastases. Scale bar represents 100 microns. (F) Tumor growth in the primary site following the transplantation of MCF7 control cells and MCF7 expressing KLF17 shRNA in mammary fat pad. There is no significant difference between two groups (p=0.775, t-test, n=10). (G) Transplantation of human breast cancer MCF7 cells stably expressing KLF17 shRNA in mammary fat pad leads to lung metastasis whereas MCF7 cells expressing a non-target shRNA did not. (H) Histology analysis of metastasis in the lung following the transplantation of MCF7 cells expressing KLF17 shRNA. Arrow indicates micrometastases. Scale bar represents 50 microns.
Figure 2
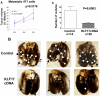
(A) The volumes of primary tumors were measured following the transplantation of 4T1 cells stably overexpressing KLF17 or a vector control in mammary fat pad. There is no statistical difference in primary tumor growth between these two groups (t-test, n=14 for the control group, n=20 for the KLF17 group). (B and C) The metastastic nodules on the surface of the lungs (B) were counted following the transplantation of 4T1 cells stably overexpressing KLF17 or a vector control in mammary fat pad. Transplantation of 4T1 cells overexpressing KLF17 leads to significant less lung metastases compared to the control (t-test) (C).
Figure 3
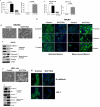
Suppression of KLF17 expression promotes tumor cell migration, invasion and EMT. (A and B) Breast cancer 168FARN and MCF7 cells stably expressing KLF17 shRNA alone or KLF17 shRNA and KLF17 cDNA were subjected to migration (A) and invasion (B) assays. The suppression of KLF17 expression in these cells leads to significant increase of cell migration and invasion. Data represent mean and s.d. (triplicates, t-test). The expression of KLF17 cDNA with no shRNA target site reverses the migratory and invasive phenotype induced by KLF17 knockdown. (C-H) The knockdown of KLF17 expression in NMuMG and HMEL cells causes epithelial-mesenchymal transition (EMT). (C, F) NMuMG and HMEL cells stably expressing KLF17 shRNA displayed spindle-like, fibroblastic morphology (20X magnification). (D, G) Immunoblots of epithelial and mesenchymal markers in NMuMG-KLF17Kd and HMEL-KLF17Kd cells show the inhibition of the expression of epithelial markers E-cadherin, ZO-1 and β-catenin and the increase of the expression of mesenchymal markers fibronectin, vimentin and N-cadherin; (E, H) Immunostaining of NMuMG-KLF17Kd and HMEL-KLF17Kd cells using antibodies shows the loss of epithelial markers in cell-cell contacts. Scale bar represents 50 microns.
Figure 4
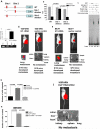
KLF17 directly binds to the promoter of Id1 and suppresses its expression. (A) Luciferase reporter constructs contain the Id-1 promoter with two potential KLF17 binding sites upstream of a luciferase gene, or Id1 promoter with the deletion of one potential binding site 1 or Id1 promoter with the deletion of potential binding site 2. Red indicates two potential binding sites (site 1: -2127 to -2110 and site 2: -1327 to -1316). (B) 168FARN cells were co-transfected with KLF17 cDNA, a control luciferase vector or luciferase reporter plasmids shown in (A) and a Renilla luciferase as a normalizing control. Luciferase activity was measured 48 hours following the transfection. The percentage of luciferase activity was determined by the activity of promoter reporter over the control luciferase vector. Data represent mean and s.d. (triplicates, t-test). (C) EMSA assay was performed using the recombinant GST-KLF17 protein and the isotope-labeled DNA probe comprising site 1. (D) Chromatin immunoprecipitation (ChIP) was performed using KLF17 antibody, acetyl-H3 antibody or control IgG. Id1 promoter region where KLF17 binds showed a significant enrichment following immunoprecipitation by KLF17 antibody. Data represent mean and s.d. (triplicates, t-test). PCR products following ChIP were run on an ethidium-stained gel. Anti-acetyl H3 antibody was used as a positive control. (E) Transplantation of 168FARN cells stably expressing mouse Id1 cDNA in mammary fat pad leads to lung metastasis. (F) Transplantation of human breast cancer MCF7 cells stably expressing human Id1 cDNA in the mammary fat pad of SCID mice leads to lung metastasis. (G-H) 168FARN cells stably expressing a KLF17 shRNA; or KLF17 and Id1 shRNAs; or a control non-target shRNA were subjected to migration (G), invasion (H) and metastasis (I) assays. Data represent mean and s.d. (triplicates, t-test). Knockdown of KLF17 significantly increase the migration, invasion and metastasis in 168FARN. Knockdown of both KLF17 and its target gene Id1 abrogates the migratory, invasive and metastasis phenotypes induced by KLF17 knockdown.
Figure 5
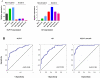
KLF17 and Id1 expression in breast cancer cell lines and their predictive value of lymph node metastasis in human primary breast cancer samples. (A) KLF17 and Id1 expression in human breast cancer cells was determined by qRT-PCR. KLF17 expression is higher in non-invasive MCF7, MDA-MB-361 and CAMA cells, but lower in invasive MDA-MB-435, MDA-MB-436, MDA-MB-453 and MDA-MB-231 cells. Id1 expression is higher in invasive cells but lower in non-invasive cells. Pearson correlation analysis indicated that the expression of KLF17 and Id-1 was inversely correlated (r=-0.75, p=0.05). (B) KLF17 and Id1 expression are predictive markers of lymph node metastasis in human primary breast cancer samples. The receiver operating characteristic curves for KLF17 (left), Id-1 (middle) and the bivariate logistic regression model for KLF17 and Id1 (right).
Similar articles
- Tumor-suppressive p53 Signaling Empowers Metastatic Inhibitor KLF17-dependent Transcription to Overcome Tumorigenesis in Non-small Cell Lung Cancer.
Ali A, Bhatti MZ, Shah AS, Duong HQ, Alkreathy HM, Mohammad SF, Khan RA, Ahmad A. Ali A, et al. J Biol Chem. 2015 Aug 28;290(35):21336-51. doi: 10.1074/jbc.M114.635730. Epub 2015 Apr 24. J Biol Chem. 2015. PMID: 25911104 Free PMC article. - Gain-of-function of mutant p53: mutant p53 enhances cancer progression by inhibiting KLF17 expression in invasive breast carcinoma cells.
Ali A, Shah AS, Ahmad A. Ali A, et al. Cancer Lett. 2014 Nov 1;354(1):87-96. doi: 10.1016/j.canlet.2014.07.045. Epub 2014 Aug 8. Cancer Lett. 2014. PMID: 25111898 - DJ-1 upregulates breast cancer cell invasion by repressing KLF17 expression.
Ismail IA, Kang HS, Lee HJ, Kim JK, Hong SH. Ismail IA, et al. Br J Cancer. 2014 Mar 4;110(5):1298-306. doi: 10.1038/bjc.2014.40. Epub 2014 Feb 6. Br J Cancer. 2014. PMID: 24504364 Free PMC article. - Krüppel-like factor 17, a novel tumor suppressor: its low expression is involved in cancer metastasis.
Zhou S, Tang X, Tang F. Zhou S, et al. Tumour Biol. 2016 Feb;37(2):1505-13. doi: 10.1007/s13277-015-4588-3. Epub 2015 Dec 12. Tumour Biol. 2016. PMID: 26662959 Free PMC article. Review. - Transcriptional regulation of metastatic [Id]entity by KLF17.
Iwanicki MP, Brugge JS. Iwanicki MP, et al. Genome Biol. 2009;10(11):244. doi: 10.1186/gb-2009-10-11-244. Epub 2009 Nov 30. Genome Biol. 2009. PMID: 19951400 Free PMC article. Review.
Cited by
- Transcriptional control of cancer metastasis.
Ell B, Kang Y. Ell B, et al. Trends Cell Biol. 2013 Dec;23(12):603-11. doi: 10.1016/j.tcb.2013.06.001. Epub 2013 Jul 6. Trends Cell Biol. 2013. PMID: 23838335 Free PMC article. Review. - A methodological approach to unravel organ-specific breast cancer metastasis.
Nola S, Sin S, Bonin F, Lidereau R, Driouch K. Nola S, et al. J Mammary Gland Biol Neoplasia. 2012 Jun;17(2):135-45. doi: 10.1007/s10911-012-9256-2. Epub 2012 May 25. J Mammary Gland Biol Neoplasia. 2012. PMID: 22628182 Review. - KLF6-SV1 drives breast cancer metastasis and is associated with poor survival.
Hatami R, Sieuwerts AM, Izadmehr S, Yao Z, Qiao RF, Papa L, Look MP, Smid M, Ohlssen J, Levine AC, Germain D, Burstein D, Kirschenbaum A, DiFeo A, Foekens JA, Narla G. Hatami R, et al. Sci Transl Med. 2013 Jan 23;5(169):169ra12. doi: 10.1126/scitranslmed.3004688. Sci Transl Med. 2013. PMID: 23345610 Free PMC article. - Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression.
Xiang X, Deng Z, Zhuang X, Ju S, Mu J, Jiang H, Zhang L, Yan J, Miller D, Zhang HG. Xiang X, et al. PLoS One. 2012;7(12):e50781. doi: 10.1371/journal.pone.0050781. Epub 2012 Dec 17. PLoS One. 2012. PMID: 23284647 Free PMC article. - GATA4 is upregulated in nasopharyngeal cancer and facilitates epithelial-mesenchymal transition and metastasis through regulation of SLUG.
Zhou Y, Chang H, Yang B. Zhou Y, et al. Exp Ther Med. 2018 Dec;16(6):5318-5326. doi: 10.3892/etm.2018.6826. Epub 2018 Oct 3. Exp Ther Med. 2018. PMID: 30542490 Free PMC article.
References
- Gupta GP, Massague J. Cancer Metastasis: Building a framework. Cell. 2006;127:679–695. - PubMed
- Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nature Rev. Med. 2006;12:895–904. - PubMed
- Fidler IJ. The pathogenesis of cancer metastasis: the “seed and soil” hypothesis revisited. Nature Rev. Cancer. 2003;3:1–6. - PubMed
- Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nature Rev. Cancer. 2003;3:55–63. - PubMed
- Yang J, Mani SA, Donaher JL, Ramaswarmy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA010815/CA/NCI NIH HHS/United States
- R01 CA148759/CA/NCI NIH HHS/United States
- R21 NS059478/NS/NINDS NIH HHS/United States
- P30 CA10815/CA/NCI NIH HHS/United States
- P50 CA083638/CA/NCI NIH HHS/United States
- P50-CA83638/CA/NCI NIH HHS/United States
- S10 RR024693/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases