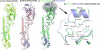Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957 - PubMed (original) (raw)
Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957
Junfeng Liu et al. Proc Natl Acad Sci U S A. 2009.
Abstract
The viruses that caused the three influenza pandemics of the twentieth century in 1918, 1957, and 1968 had distinct hemagglutinin receptor binding glycoproteins that had evolved the capacity to recognize human cell receptors. We have determined the structure of the H2 hemagglutinin from the second pandemic, the "Asian Influenza" of 1957. We compare it with the 1918 "Spanish Influenza" hemagglutinin, H1, and the 1968 "Hong Kong Influenza" hemagglutinin, H3, and show that despite its close overall structural similarity to H1, and its more distant relationship to H3, the H2 receptor binding site is closely related to that of H3 hemagglutinin. By analyzing hemagglutinins of potential H2 avian precursors of the pandemic virus, we show that the human receptor can be bound by avian hemagglutinins that lack the human-specific mutations of H2 and H3 pandemic viruses, Gln-226Leu, and Gly-228Ser. We show how Gln-226 in the avian H2 receptor binding site, together with Asn-186, form hydrogen bond networks through bound water molecules to mediate binding to human receptor. We show that the human receptor adopts a very similar conformation in both human and avian hemagglutinin-receptor complexes. We also show that Leu-226 in the receptor binding site of human virus hemagglutinins creates a hydrophobic environment near the Sia-1-Gal-2 glycosidic linkage that favors binding of the human receptor and is unfavorable for avian receptor binding. We consider the significance for the development of pandemics, of the existence of avian viruses that can bind to both avian and human receptors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Ribbons representation of different H2 HA monomers and receptor binding sites. (A) Superposition of the monomers of two human H2 HAs: A/Singapore/1/57 and A/Japan/305/57 colored green and yellow respectively. (B) Three avian H2 HAs: A/ck/New York/29878/91 colored gray, A/dk/Ontario/77 colored in blue and A/ck/potsdam/4705/84 colored orange red. (C) Overlap of monomers of a human H2 HA colored in green and avian H2 HA colored in blue. The region highlighted by the gray ellipse at the top of the panel shows the receptor binding domain, an expanded version of which is shown in (D). Conserved residues such as Tyr-98, Ser-136, Trp-153, and His-183 are shown in stick representation together with other residues important in receptor binding specificity such as Asn-186, Glu-190, and Leu-194, as well as the Gln/Leu-226, Gly/Ser-228 pair.
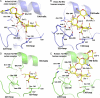
Fig. 2.
Interactions of an avian H2 HA (upper panels) and a human H2 HA (lower panels) with avian and human receptor analogues. The three secondary structure elements of the binding site, the 130- and 220-loops and the 190-helix are labeled in this backbone representation together with some selected side chains in stick representation. The broken lines indicate potential hydrogen bond interaction. In all four panels, the sialosaccharides are colored yellow for carbon atoms, blue for nitrogen, and red for oxygen, water molecules are indicated by red spheres. A/dk/Ontario/77 H2 HA, colored blue, in complex with avian receptor, LSTa, (A) and human receptor, LSTc. (B). A/Singapore/1/57 H2 HA, colored in green, in complex with human receptor (C) and avian receptor (D). The black arrows in A, B, and C indicate that for the two human receptor complexes the Sia-1/Gal-2 linkage adopts a cis conformation, whereas for the avian complex it adopts a trans conformation.
Fig. 3.
Overlap of the receptor binding domains of H1, H2, and H3 HAs in complexes with receptor analogues. (A) The overlapped receptor binding sites of human HAs for H1 A/S.Carolina/1918, (blue), H2 A/Singapore/1/57, (yellow) and H3 A/Aichi/2/68, (22) (gray) in complex with human receptor analogue, LSTc. (B) The receptor binding domains of avian HAs for H1, A/dk/Alberta/76 (blue), H2, A/dk/Ontario/77 (yellow), and H3, A/dk/Ukraine/63 (25) (gray) in complex with human receptor analogue. The sialopentasaccharides are colored according to the HAs to which they are bound and some of the side-chains discussed in the text are shown in stick representation.
Similar articles
- X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus.
Ha Y, Stevens DJ, Skehel JJ, Wiley DC. Ha Y, et al. Virology. 2003 May 10;309(2):209-18. doi: 10.1016/s0042-6822(03)00068-0. Virology. 2003. PMID: 12758169 - X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs.
Ha Y, Stevens DJ, Skehel JJ, Wiley DC. Ha Y, et al. Proc Natl Acad Sci U S A. 2001 Sep 25;98(20):11181-6. doi: 10.1073/pnas.201401198. Epub 2001 Sep 18. Proc Natl Acad Sci U S A. 2001. PMID: 11562490 Free PMC article. - Adaptation of influenza viruses to human airway receptors.
Thompson AJ, Paulson JC. Thompson AJ, et al. J Biol Chem. 2021 Jan-Jun;296:100017. doi: 10.1074/jbc.REV120.013309. Epub 2020 Nov 22. J Biol Chem. 2021. PMID: 33144323 Free PMC article. Review. - Receptor binding by an H7N9 influenza virus from humans.
Xiong X, Martin SR, Haire LF, Wharton SA, Daniels RS, Bennett MS, McCauley JW, Collins PJ, Walker PA, Skehel JJ, Gamblin SJ. Xiong X, et al. Nature. 2013 Jul 25;499(7459):496-9. doi: 10.1038/nature12372. Nature. 2013. PMID: 23787694 - Enabling the 'host jump': structural determinants of receptor-binding specificity in influenza A viruses.
Shi Y, Wu Y, Zhang W, Qi J, Gao GF. Shi Y, et al. Nat Rev Microbiol. 2014 Dec;12(12):822-31. doi: 10.1038/nrmicro3362. Epub 2014 Nov 10. Nat Rev Microbiol. 2014. PMID: 25383601 Review.
Cited by
- Molecular Markers and Mechanisms of Influenza A Virus Cross-Species Transmission and New Host Adaptation.
Guo X, Zhou Y, Yan H, An Q, Liang C, Liu L, Qian J. Guo X, et al. Viruses. 2024 May 30;16(6):883. doi: 10.3390/v16060883. Viruses. 2024. PMID: 38932174 Free PMC article. Review. - The Influenza A Virus Replication Cycle: A Comprehensive Review.
Carter T, Iqbal M. Carter T, et al. Viruses. 2024 Feb 19;16(2):316. doi: 10.3390/v16020316. Viruses. 2024. PMID: 38400091 Free PMC article. Review. - Has avian influenza virus H9 originated from a bat source?
Karamendin K, Kydyrmanov A, Fereidouni S. Karamendin K, et al. Front Vet Sci. 2024 Jan 8;10:1332886. doi: 10.3389/fvets.2023.1332886. eCollection 2023. Front Vet Sci. 2024. PMID: 38260204 Free PMC article. - Global distribution, receptor binding, and cross-species transmission of H6 influenza viruses: risks and implications for humans.
Yan Z, Li Y, Huang S, Wen F. Yan Z, et al. J Virol. 2023 Nov 30;97(11):e0137023. doi: 10.1128/jvi.01370-23. Epub 2023 Oct 25. J Virol. 2023. PMID: 37877722 Free PMC article. Review. - Genetic characterization of a new candidate hemagglutinin subtype of influenza A viruses.
Fereidouni S, Starick E, Karamendin K, Genova CD, Scott SD, Khan Y, Harder T, Kydyrmanov A. Fereidouni S, et al. Emerg Microbes Infect. 2023 Dec;12(2):2225645. doi: 10.1080/22221751.2023.2225645. Emerg Microbes Infect. 2023. PMID: 37335000 Free PMC article.
References
- Lim KA, Smith A, Hale JH, Glass J. Influenza outbreak in Singapore. Lancet. 1957;273:791–796. - PubMed
- Meyer HM, Jr, Hilleman MR, Miesse ML, Crawford IP, Bankhead AS. New antigenic variant in Far East influenza epidemic, 1957. Proc Soc Exp Biol Med. 1957;95:609–616. - PubMed
- Chu CM. The etiology and epidemiology of influenza: An analysis of the 1957 epidemic. J Hyg Epidemiol Microbiol Immunol. 1958;2:1–8. - PubMed
- Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical