Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation - PubMed (original) (raw)
Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation
Min-Ji Kang et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Mutations in the rhodopsin gene that disrupt the encoded protein's folding properties are a major cause of autosomal dominant retinitis pigmentosa (ADRP). This disease is faithfully modeled in Drosophila where similar mutations in the ninaE gene, encoding rhodopsin-1 (Rh-1), cause ER stress and dominantly trigger age-related retinal degeneration. In addition, mutant flies bearing certain ninaE alleles have dramatically reduced Rh-1 protein levels, but the underlying mechanism for this reduction and significance of its contribution to the ADRP phenotype remains unclear. To address this question, we specifically analyzed the role of Drosophila genes homologous to the known yeast and animal regulators of the ER-associated degradation (ERAD) pathway, a process that reduces levels of misfolded proteins in the ER through proteasomal degradation. We found that loss-of-function of these putative ERAD factors resulted in increased levels of Rh-1 in ninaE mutant flies. Conversely, in an ER stress assay where mutant or wild-type Rh-1 were overexpressed in developing imaginal discs beyond the ER protein folding capacity of those cells, co-expression of certain ERAD factors was sufficient to reduce Rh-1 protein levels and to completely suppress ER stress reporter activation. Significantly, those ERAD factors that specifically reduced misfolded Rh-1 in the imaginal disc assay also delayed age-related retinal degeneration caused by an endogenous ninaE allele, indicating that ERAD acts as a protective mechanism against retinal degeneration in the Drosophila model for ADRP. These results suggest that manipulation of ERAD may serve as a powerful therapeutic strategy against a number of diseases associated with ER stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
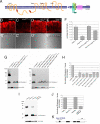
Loss of ERAD regulators partially restores the level of Rh-1 in the retina of _ninaEG69D_−/+ flies. (A) Schematic diagrams of the predicted membrane topology and domain structure of ERAD components through EMBnet-CH. (B–E) Horizontal retina cryosections stained with anti-Rh-1 antibody in 1- to 3-day-old flies. (B) A control ninaE +/+ retina. _ninaEG69D_−/+ retina (C) show low Rh-1 labeling, while Rh-1 levels recover when Hrd1 (D) or Herp (E) are knocked down in this genetic background. In all RNAi experiments, dicer2 (drc2) were co-expressed to enhance the efficiency of RNAi knockdown. (F) The average intensity of anti-Rh-1 labeling (n = 3), as quantified through the Image J. Error bars, ± SEM. (G and I) A Western blot of Drosophila adult head extracts of the indicated genotypes probed for Rh-1 (upper gel) and anti-profilin (as a loading control; lower gel). (H and J) Quantification of the average normalized Rh-1 band intensity as shown in (G and I) (average of n = 3; Der-1, average of n = 2), with the value from wild-type of Rh-1+/+ heads extracts set at 100%. (K) The structure of the Herp genomic locus. In the _Herp_G13463, an EP-element is inserted within its protein-coding region. Genotypes: Rh1-Gal4;;ninaE+/+ (B and _B_′), Rh1-Gal4;;ninaEG69D/+ (C and _C_′), Rh1-Gal4;UAS-Dcr2/+;UAS-Hrd1-IR/ninaEG69D (D and _D_′), Rh1-Gal4;UAS-Dcr2/+;UAS-Herp-IR/ninaEG69D (E and _E_′), (G, lanes 1 and 5) Rh1-Gal4;;ninaE+/+, (G, lanes 2 and 6) Rh1-Gal4;;ninaEG69D/+, (G, lane 3) Rh1-Gal4;UAS-Dcr2/+;UAS-Hrd1-IR/ninaEG69D, (G, lane 4) Rh1-Gal4;UAS-Hrd3-IR/+;UAS-Dcr2/ninaEG69D, (G, lane 7) Rh1-Gal4;UAS-Herp-IR/+;UAS-Dcr2/ninaEG69D, (G, lane 8) Rh1-Gal4;UAS-Dcr2/+;UAS-Herp-IR/ninaEG69D, (G, lane 9) Rh1-Gal4;UAS-Dcr2/+;UAS-Der-1-IR(#1)/ninaEG69D, (G, lane 10) Rh1-Gal4;UAS-Dcr2/+;UAS-Der-1-IR(#2)/ninaEG69D, (I, lane 1) CantonS, (I, lane 2) ninaEG69D/+, (I, lane 3) HerpG13463/HerpG13463, (I, lane 4) HerpG13463/HerpG13463;ninaEG69D/+.
Fig. 2.
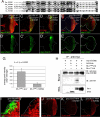
ER stress caused by Rh-1 misexpression is strongly suppressed by Drosophila Hrd1. (A–D) Representative images of eye imaginal discs expressing Rh-1WT (A) or Rh-1G69D (C) alone, or together with Hrd1 (B and D). Anti-Rh-1 antibody labeling is in red. E–I Hrd1 co-expression abolished ER stress caused by wild-type or mutant Rh-1 misexpression, as determined by the ER stress marker, xbp1-EGFP (green). Shown are representative discs expressing xbp1-EGFP alone (E), or together with indicated genes. (J) Co-immunoprecipitation assays between Hrd1 and Rh-1 in 293T cells. Hrd1 was tagged with the myc epitope, while Rh-1 was tagged with HA. HA-Drob-1 is a membrane protein used as a negative control. [Scale bars, 100 μm (A).] Genotypes: gmr-Gal4/+;_UAS-Rh-1_WT/+ (A), gmr-Gal4/UAS-Hrd1;_UAS-Rh-1_WT/+ (B), gmr-Gal4/+;UAS-Rh-1G69D/+ (C), gmr-Gal4/UAS-Hrd1;UAS-Rh-1G69D/+ (D), gmr-Gal4/+;UAS-xbp1-EGFP/+ (E), gmr-Gal4/UAS-lacZ;_UAS-Rh-1_WT/UAS-xbp1-EGFP (F), gmr-Gal4/UAS-Hrd1;_UAS-Rh-1_WT/UAS-xbp1-EGFP (G), gmr-Gal4/UAS-lacZ;UAS- Rh-1G69D/UAS-xbp1-EGFP (H), and gmr-Gal4/UAS-Hrd1;UAS-Rh-1G69D/UAS-xbp1-EGFP (I).
Fig. 3.
Drosophila EDEM2 overexpression reduces mutant, but not wild-type Rh-1 levels. (A) The amino acid sequence alignment between EDEM family proteins of Drosophila and humans, generated with the ClustalW algorithm. Dark shading indicates identity, whereas light shading indicates similarity. Solid lines indicate the catalytic domain of class I mannosidase (Glycosyl hydrolase family 47). The sequences shown, with their corresponding NCBI database accession numbers, are as follows: D. melanogaster (Dm) EDEM1 (NP_726777); D. melanogaster (Dm) EDEM2 (AAF53255); human (Hs) EDEM1 (AAH19088); human (Hs) EDEM2 (NP_060687); human (Hs) EDEM3 (NP_079467). (B–D) The effect of Drosophila EDEM1 and EDEM2 on misexpressed Rh-1WT. Shown are representative discs expressing Rh-1WT, together with lacZ (B), EDEM1 (C), or EDEM2 (D), or expressing Rh-1G69D with lacZ (E) or EDEM2 (F). The degree of ER stress is assessed through xbp1-EGFP activation (_B_′–_F_′, green). (G) Quantification of the xbp1-EGFP activation levels as shown in (E and F). Error bars, ± SEM. (H) EDEM2 physically interacts with Rh-1G69D in S2 cells. EDEM2 was immunoprecipitated through its myc-tag, and its interaction partners were detected through their HA-epitopes. (I and _I_′) A disc expressing EDEM2 (green) in a flip-out mosaic clone shows reduced levels of Rh-1G69D (red). (J–L) Both of EDEM1 and EDEM2 effectively downregulated the level of alpha 1-antitrypsinNHK (A1AT NHK, in red). (Scale bars, 100 μm (B) and 10 μm (I).) Genotypes: gmr-Gal4/UAS-lacZ;_UAS-Rh-1_WT/UAS-xbp1-EGFP (B and _B_′), gmr-Gal4/+;_UAS-Rh-1_WT, UAS-EDEM1/UAS-xbp1-EGFP (C and _C_′), gmr-Gal4/UAS-EDEM2;_UAS-Rh-1_WT/UAS-xbp1-EGFP (D and _D_′), gmr-Gal4/UAS-lacZ;UAS-Rh-1G69D/UAS-xbp1-EGFP (E and _E_′), gmr-Gal4/UAS-EDEM2;UAS-Rh-1G69D/UAS-xbp1-EGFP (F and _F_′), hs-flp;UAS-EDEM2/+;_tub_>_GFP_>Gal4/gmr-Rh-1G69D (I and _I_′), gmr-Gal4/UAS-alpha1-antitrypsinNHK;+/+ (J), gmr-Gal4/UAS-alpha1-antitrypsinNHK;UAS-EDEM1/+ (K), and gmr-Gal4/UAS-alpha1-antitrypsinNHK, UAS-EDEM2;+/+ (L).
Fig. 4.
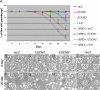
Drosophila Hrd1 and EDEM2 overexpression suppresses late onset retinal degeneration of _ninaEG69D_−/+ flies. (A) Quantification of the extent of retinal degeneration through the pseudopupil assay. For each genenotype, the graph shows the percentage of flies with intact pseudopupils (average of four independent crosses). Specifically, lacZ, EDEM1, EDEM2 or Hrd1 were overexpressed through the Rh1-Gal4 driver in ninaE mutant or wild type backgrounds. Overexpression of Hrd1 and EDEM2 delay the course of retinal degeneration of _ninaEG69D_−/+ flies. (B–I) Representative images of adult retina overexpressing designated ERAD factors in the _ninaEG69D_−/+ background. (B–E) At day 0 after eclosion, retina of all genotypes have regular array of ommatidia. (F–I) Retinal sections of indicated genotypes at day 20. The degree of ommatidial disarray is suppressed by overexpressing EDEM2 or Hrd1 (H and I), but not by lacZ (F) or EDEM1 (G).
Similar articles
- Characterization of two dominant alleles of the major rhodopsin-encoding gene ninaE in Drosophila.
Mitra A, Chinchore Y, Kinser R, Dolph PJ. Mitra A, et al. Mol Vis. 2011;17:3224-33. Epub 2011 Dec 14. Mol Vis. 2011. PMID: 22194648 Free PMC article. - Inactivation of VCP/ter94 suppresses retinal pathology caused by misfolded rhodopsin in Drosophila.
Griciuc A, Aron L, Roux MJ, Klein R, Giangrande A, Ueffing M. Griciuc A, et al. PLoS Genet. 2010 Aug 26;6(8):e1001075. doi: 10.1371/journal.pgen.1001075. PLoS Genet. 2010. PMID: 20865169 Free PMC article. - Drosophila arf72A acts as an essential regulator of endoplasmic reticulum quality control and suppresses autosomal-dominant retinopathy.
Lee J, Lee J, Ju BG. Lee J, et al. Int J Biochem Cell Biol. 2011 Sep;43(9):1392-401. doi: 10.1016/j.biocel.2011.06.004. Epub 2011 Jun 12. Int J Biochem Cell Biol. 2011. PMID: 21693198 - Molecular genetics of retinal degeneration: A Drosophila perspective.
Shieh BH. Shieh BH. Fly (Austin). 2011 Oct-Dec;5(4):356-68. doi: 10.4161/fly.5.4.17809. Epub 2011 Sep 7. Fly (Austin). 2011. PMID: 21897116 Free PMC article. Review.
Cited by
- ER stress-induced cell death mechanisms.
Sano R, Reed JC. Sano R, et al. Biochim Biophys Acta. 2013 Dec;1833(12):3460-3470. doi: 10.1016/j.bbamcr.2013.06.028. Epub 2013 Jul 10. Biochim Biophys Acta. 2013. PMID: 23850759 Free PMC article. Review. - Meep, a Novel Regulator of Insulin Signaling, Supports Development and Insulin Sensitivity via Maintenance of Protein Homeostasis in Drosophila melanogaster.
Pereira MT, Brock K, Musselman LP. Pereira MT, et al. G3 (Bethesda). 2020 Dec 3;10(12):4399-4410. doi: 10.1534/g3.120.401688. G3 (Bethesda). 2020. PMID: 32998936 Free PMC article. - Immune modulation by MANF promotes tissue repair and regenerative success in the retina.
Neves J, Zhu J, Sousa-Victor P, Konjikusic M, Riley R, Chew S, Qi Y, Jasper H, Lamba DA. Neves J, et al. Science. 2016 Jul 1;353(6294):aaf3646. doi: 10.1126/science.aaf3646. Science. 2016. PMID: 27365452 Free PMC article. - Protein misfolding and dysregulated protein homeostasis in autoinflammatory diseases and beyond.
Agyemang AF, Harrison SR, Siegel RM, McDermott MF. Agyemang AF, et al. Semin Immunopathol. 2015 Jul;37(4):335-47. doi: 10.1007/s00281-015-0496-2. Epub 2015 May 21. Semin Immunopathol. 2015. PMID: 25994946 Review. - Cluster-Based Analysis of Retinitis Pigmentosa Modifiers Using Drosophila Eye Size and Gene Expression Data.
Amstutz J, Khalifa A, Palu R, Jahan K. Amstutz J, et al. Genes (Basel). 2022 Feb 21;13(2):386. doi: 10.3390/genes13020386. Genes (Basel). 2022. PMID: 35205430 Free PMC article.
References
- Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86:1133–1149. - PubMed
- Dryja T, et al. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–366. - PubMed
- O'Tousa JE, et al. The Drosophila ninaE gene encodes an opsin. Cell. 1985;40:877–882. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials