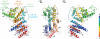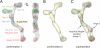Structure of a trimeric nucleoporin complex reveals alternate oligomerization states - PubMed (original) (raw)
Structure of a trimeric nucleoporin complex reveals alternate oligomerization states
Vivien Nagy et al. Proc Natl Acad Sci U S A. 2009.
Abstract
The heptameric Nup84 complex constitutes an evolutionarily conserved building block of the nuclear pore complex. Here, we present the crystal structure of the heterotrimeric Sec13 x Nup145C x Nup84 complex, the centerpiece of the heptamer, at 3.2-A resolution. Nup84 forms a U-shaped alpha-helical solenoid domain, topologically similar to two other members of the heptamer, Nup145C and Nup85. The interaction between Nup84 and Nup145C is mediated via a hydrophobic interface located in the kink regions of the two solenoids that is reinforced by additional interactions of two long Nup84 loops. The Nup84 binding site partially overlaps with the homo-dimerization interface of Nup145C, suggesting competing binding events. Fitting of the elongated Z-shaped heterotrimer into electron microscopy (EM) envelopes of the heptamer indicates that structural changes occur at the Nup145C x Nup84 interface. Docking the crystal structures of all heptamer components into the EM envelope constitutes a major advance toward the completion of the structural characterization of the Nup84 complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Structure of the S. cerevisiae Sec13·Nup145C·Nup84 NTD complex. (A) Schematic representation of the heptameric complex and the approximate localization of its seven nups (21). (B) Domain structures of Sec13, Nup145C, and Nup84. For Sec13, the six WD40 repeats (orange) are indicated. For Nup145C, the unstructured N-terminal region (gray), the domain invasion motif (DIM) (green), the αB-αC connector (C) (red), the α-helical domain (blue), and the C-terminal α-helical region (pink) are indicated. For Nup84, the N-terminal domain (NTD) and C-terminal domain (CTD) are indicated. The residue numbering is shown below and the bars above the domain structures mark the crystallized fragments of the three proteins. (C) Structure of Sec13·Nup145C·Nup84 NTD in ribbon representation, colored as in panel B. A 90°-rotated view is shown on the right. (D) Schematic representation of the Sec13·Nup145C·Nup84 NTD heterotrimer.
Fig. 2.
The Nup84 α-helical domain. Ribbon representation of the Nup84 NTD is shown in rainbow colors along the polypeptide chain from the N- to the C- terminus. The four loops that participate in the Nup145C·Nup84 interaction are indicated.
Fig. 3.
Surface properties of the Nup84 NTD. The surface orientations are identical in all columns. A black line encircles the Nup145C interaction surface. (A) Surface rendition of the Nup84 NTD. The Nup145C contact surface is colored in blue, while the remaining surface is colored in yellow. As a reference, a surface rendition of the heterotrimer is shown to the left, colored according to Fig. 1_C_. (B) Surface representation colored according to a multispecies sequence alignment, ranging from 60% similarity (white) to 100% identity (red) (
Fig. S4
). (C) Surface rendition colored according to the electrostatic potential, ranging from −10 kBT/e (red) to + 10 kBT/e (blue). Note the two conserved hydrophobic patches located toward the periphery of the extended Nup145C-interacting surface.
Fig. 4.
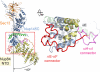
Interaction of the Nup84 NTD with the Nup145C solenoid domain. The Sec13·Nup145C·Nup84 NTD heterotrimer is shown in ribbon representation, colored according to Fig. 1_C_. The kink regions of the two solenoids interact in a head-to-head fashion. The Nup84 NTD protrudes with an approximate 40° angle from the Nup145C U-shaped solenoid. The inset marks the Nup145C·Nup84 interface that is illustrated in detail on the right. For clarity, the interface shown on the right is rotated by 90°. For Nup145C, the solenoid subdomain (blue) and helix αE (green) are indicated. For Nup84, the interface helices (yellow), as well as the long αE-αF (red) and αH-αI (magenta) connectors that mediate the interaction with Nup145C are indicated.
Fig. 5.
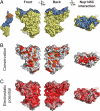
Binding promiscuity of Nup145C. (A) Surface rendition of the Sec13·Nup145C nucleoporin pair derived from the Sec13·Nup145C hetero-octamer (Nup145C·Nup145C homo-dimerization) and the Sec13·Nup145C·Nup84 NTD heterotrimer (Nup145C·Nup84 NTD hetero-dimerization). The Nup145C homo-dimerization and hetero-dimerization surfaces are colored in green and yellow, respectively. The Sec13 and the remaining Nup145C surfaces are colored in orange and blue, respectively. (B) 90°-rotated views of the Sec13·Nup145C pair colored according to panel A (top), to a multispecies sequence alignment, ranging from 60% similarity (white) to 100% identity (red) (26) (middle), and to the electrostatic potential, from −10 kBT/e (red) to +10 kBT/e (blue). The orientation of all surface representations is identical in each column. As a reference, black lines encircle the Nup145C homo-dimerization and Nup84-interaction surfaces.
Fig. 6.
Protein arrangement within the heptameric complex. (A) Docking of crystal structures into the EM envelope of the heptameric Nup84 complex. A 90°-rotated view is shown on the right. The approximate 40° angle by which the Nup84 NTD protrudes from the Sec13·Nup145 nucleoporin pair nicely follows one of the two kink regions of the heptamer stem. (B) EM envelope of the second reconstructed conformation of the heptamer in which the two hinge regions are completely extended, forming an almost entirely straight stem. (C) Superposition of the two determined heptamer conformations. The kink region at the Nup145C·Nup84 interface is indicated and was used for the structural alignment, showing that this interface corresponds to a hinge in the heptamer stem.
Similar articles
- Characterization of the membrane-coating Nup84 complex: paradigm for the nuclear pore complex structure.
Debler EW, Hsia KC, Nagy V, Seo HS, Hoelz A. Debler EW, et al. Nucleus. 2010 Mar-Apr;1(2):150-7. doi: 10.4161/nucl.1.2.11120. Epub 2010 Jan 3. Nucleus. 2010. PMID: 21326946 Free PMC article. Review. - Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice.
Brohawn SG, Schwartz TU. Brohawn SG, et al. Nat Struct Mol Biol. 2009 Nov;16(11):1173-7. doi: 10.1038/nsmb.1713. Epub 2009 Oct 25. Nat Struct Mol Biol. 2009. PMID: 19855394 Free PMC article. - The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture.
Leksa NC, Brohawn SG, Schwartz TU. Leksa NC, et al. Structure. 2009 Aug 12;17(8):1082-91. doi: 10.1016/j.str.2009.06.003. Epub 2009 Jul 2. Structure. 2009. PMID: 19576787 Free PMC article. - Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex.
Seo HS, Ma Y, Debler EW, Wacker D, Kutik S, Blobel G, Hoelz A. Seo HS, et al. Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14281-6. doi: 10.1073/pnas.0907453106. Epub 2009 Aug 11. Proc Natl Acad Sci U S A. 2009. PMID: 19706512 Free PMC article. - The structure of the nuclear pore complex.
Hoelz A, Debler EW, Blobel G. Hoelz A, et al. Annu Rev Biochem. 2011;80:613-43. doi: 10.1146/annurev-biochem-060109-151030. Annu Rev Biochem. 2011. PMID: 21495847 Review.
Cited by
- Characterization of the membrane-coating Nup84 complex: paradigm for the nuclear pore complex structure.
Debler EW, Hsia KC, Nagy V, Seo HS, Hoelz A. Debler EW, et al. Nucleus. 2010 Mar-Apr;1(2):150-7. doi: 10.4161/nucl.1.2.11120. Epub 2010 Jan 3. Nucleus. 2010. PMID: 21326946 Free PMC article. Review. - In situ structural analysis of the human nuclear pore complex.
von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, Buczak K, Mosalaganti S, Hagen W, Andres-Pons A, Lemke EA, Bork P, Antonin W, Glavy JS, Bui KH, Beck M. von Appen A, et al. Nature. 2015 Oct 1;526(7571):140-143. doi: 10.1038/nature15381. Epub 2015 Sep 23. Nature. 2015. PMID: 26416747 Free PMC article. - Toward the atomic structure of the nuclear pore complex: when top down meets bottom up.
Hoelz A, Glavy JS, Beck M. Hoelz A, et al. Nat Struct Mol Biol. 2016 Jul;23(7):624-30. doi: 10.1038/nsmb.3244. Epub 2016 Jun 6. Nat Struct Mol Biol. 2016. PMID: 27273515 Free PMC article. Review. - A jumbo problem: mapping the structure and functions of the nuclear pore complex.
Fernandez-Martinez J, Rout MP. Fernandez-Martinez J, et al. Curr Opin Cell Biol. 2012 Feb;24(1):92-9. doi: 10.1016/j.ceb.2011.12.013. Epub 2012 Feb 8. Curr Opin Cell Biol. 2012. PMID: 22321828 Free PMC article. Review. - Toward understanding the structure of the vertebrate nuclear pore complex.
Beck M, Glavy JS. Beck M, et al. Nucleus. 2014 Mar-Apr;5(2):119-23. doi: 10.4161/nucl.28739. Epub 2014 Apr 3. Nucleus. 2014. PMID: 24699243 Free PMC article. Review.
References
- Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. - PubMed
- Hoelz A, Blobel G. Cell biology: Popping out of the nucleus. Nature. 2004;432:815–816. - PubMed
- Debler EW, Blobel G, Hoelz A. Nuclear transport comes full circle. Nat Struct Mol Biol. 2009;5:457–459. - PubMed
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases