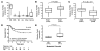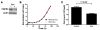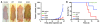Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models - PubMed (original) (raw)
. 2009 Nov;119(11):3395-407.
doi: 10.1172/JCI39703. Epub 2009 Oct 5.
Adam T Cheuk, Patricia S Tsang, Joon-Yong Chung, Young K Song, Krupa Desai, Yanlin Yu, Qing-Rong Chen, Kushal Shah, Victoria Youngblood, Jun Fang, Su Young Kim, Choh Yeung, Lee J Helman, Arnulfo Mendoza, Vu Ngo, Louis M Staudt, Jun S Wei, Chand Khanna, Daniel Catchpoole, Stephen J Qualman, Stephen M Hewitt, Glenn Merlino, Stephen J Chanock, Javed Khan
Affiliations
- PMID: 19809159
- PMCID: PMC2769177
- DOI: 10.1172/JCI39703
Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models
James G Taylor 6th et al. J Clin Invest. 2009 Nov.
Abstract
Rhabdomyosarcoma (RMS) is a childhood cancer originating from skeletal muscle, and patient survival is poor in the presence of metastatic disease. Few determinants that regulate metastasis development have been identified. The receptor tyrosine kinase FGFR4 is highly expressed in RMS tissue, suggesting a role in tumorigenesis, although its functional importance has not been defined. Here, we report the identification of mutations in FGFR4 in human RMS tumors that lead to its activation and present evidence that it functions as an oncogene in RMS. Higher FGFR4 expression in RMS tumors was associated with advanced-stage cancer and poor survival, while FGFR4 knockdown in a human RMS cell line reduced tumor growth and experimental lung metastases when the cells were transplanted into mice. Moreover, 6 FGFR4 tyrosine kinase domain mutations were found among 7 of 94 (7.5%) primary human RMS tumors. The mutants K535 and E550 increased autophosphorylation, Stat3 signaling, tumor proliferation, and metastatic potential when expressed in a murine RMS cell line. These mutants also transformed NIH 3T3 cells and led to an enhanced metastatic phenotype. Finally, murine RMS cell lines expressing the K535 and E550 FGFR4 mutants were substantially more susceptible to apoptosis in the presence of a pharmacologic FGFR inhibitor than the control cell lines expressing the empty vector or wild-type FGFR4. Together, our results demonstrate that mutationally activated FGFR4 acts as an oncogene, and these are what we believe to be the first known mutations in a receptor tyrosine kinase in RMS. These findings support the potential therapeutic targeting of FGFR4 in RMS.
Figures
Figure 1. High FGFR4 expression in RMS is associated with advanced stage, ARMS histology, and poor survival.
(A) Log2 median-centered expression data showed high FGFR4 expression in RMS tumors compared with other pediatric tumors and normal tissue of human tumors. EWS, Ewings sarcoma; NB, neuroblastoma; ALL, acute lymphoblastic leukemia; other_,_ other pediatric tumors; normal, normal tissue. (B) Stage 4 RMS was associated with significantly higher median FGFR4 expression than stage 1 RMS, as analyzed based on the data of Davicioni et al. (Mann-Whitney test) (6). (C) FGFR4 expression was significantly higher in ARMS compared with all other histologic RMS subtypes (Mann-Whitney test). (D) Kaplan-Meier analysis based on a cutoff of median FGFR4 expression showed that patients with higher expression (top 2 quartiles) had significantly higher mortality (than the bottom 2 quartiles; P = 0.03, log-rank test). (E) Murine RMS cell lines (derived from refs. 5, 21) of high metastatic potential had significantly higher median Fgfr4 mRNA than nonmetastatic cell lines (P = 0.01, Mann-Whitney test).
Figure 2. FGFR4 knockdown with an inducible shRNA leads to reduced in vitro growth.
(A) Western blot confirmed suppression of FGFR4 with doxycycline induction of the shRNA in RH30 cells. (B) Growth curves for RH30 cell lines with an inducible shRNA directed against FGFR4 showed no measurable difference in growth rate for up to 96 hours. (C) Cell count of RH30 cell lines with an inducible shRNA against FGFR4 treated with or without doxycycline shows significantly reduced growth (41% reduction) with prolonged culture at 13 days (Mann-Whitney test).
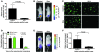
Figure 3. FGFR4 suppression leads to inhibition of in vivo growth and lung metastasis.
(A) Intramuscular injection of RH30 with inducible anti-FGFR4 shRNA resulted in significantly smaller tumors in the FGFR4-suppressed mice on day 31 using bioluminescent imaging (n = 6 mice per group, Mann-Whitney test). (B) Representative mice with intramuscular injection on day 31. The intensity of tumor cells expressing luciferase is quantified as photons/second/cm2/steridian (p/s/cm2/sr). (C) Representative IVVM images showing decreased RH30 cells in the lungs at 24 hours when FGFR4 was suppressed. (D) Quantification of IVVM early pulmonary metastases at 1 and 24 hours showed significantly fewer malignant cells remaining in the lungs with FGFR4 suppression (normalized mean values ± SEM; n = 5 mice per group; Mann-Whitney test). (E) Representative mice with intravenous injection showed decreased tumor signal in the lungs with FGFR4 suppression on day 74. (F) Intravenous injection of the same cells resulted in significantly fewer lesions in the lungs on day 74 in the FGFR4-suppressed mice (normalized for tumor growth rate by taking the ratio of pulmonary to pelvic tumor signal; n = 10 mice per group, Mann-Whitney test).
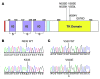
Figure 4. FGFR4 TK domain mutations in RMS.
(A) Sites of 6 missense substitutions in the FGFR4 TK domain that were identified in RMS tumors (n = 94). Amino acid boundaries for protein domains were defined by the results of a search of the NCBI Conserved Domain database (NCBI CD-Search). Red, signal peptide; blue, transmembrane domain. IG, immunoglobulin-like domain; S, disulfide bond. (B) Wild-type and homozygous mutant alleles for the N535K mutation. (C) Wild-type and mutant alleles for V550E.
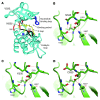
Figure 5. Structural modeling of the FGFR4 codon 535 and 550 mutations.
(A) Codon 535 and 550 mutations (red) on a model of the FGFR4 kinase domain. Mutation sites of codons 529 and 554 are also in red. The activation loop (A loop) is yellow, the catalytic loop black, and the nucleotide binding loop blue. (B) Predicted hydrogen bonds for wild-type codon 535. (C and D) Codon 535 mutations could disrupt R-group hydrogen bonds (red dashed lines) between codon 535 and residues H530 and I533. Illustrations in B–D were created with PyMOL (http://www.pymol.org).
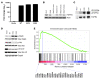
Figure 6. FGFR4 mutations promote FGFR4 autophosphorylation, STAT3 phosphorylation, and activation of cell cycle and DNA replication pathways.
(A and B) High levels of RNA (A) and protein (B) for wild-type human FGFR4, FGFR4 K535, and FGFR4 E550 after stable introduction into murine RMS cell line RMS772. Protein levels are comparable to those of human RMS cell lines RH41 and RH28. (C) Immunoblot of FGFR4 and phospho-FGFR4 after immunoprecipitation showed that the mutant forms of FGFR4 were constitutively autophosphorylated. (D) Increased total Stat3 and phospho-Stat3 were observed in FGFR4 mutant cell lines by immunoblot, while mutants also showed decreased phospho-Akt. All transductants expressing human FGFR4 also had less phospho-Erk1/2. (E) Representative GSEA showing enrichment of cell cycle genes in murine RMS cells expressing FGFR4 mutations. Gene names listed at the bottom are at the leading edge of genes ranked by expression enrichment score that also belong to the Cell Cycle KEGG gene set.
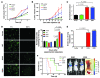
Figure 7. FGFR4 mutations accelerate growth and promote a metastasis phenotype.
(A) Growth was significantly higher for cultured mutant RMS772 cell lines (*P < 0.01 versus wild type, Mann-Whitney test). (B) FGFR4 mutants in RMS772 cells significantly enhanced in vivo growth after subcutaneous injection in nude mice (*P < 0.001 versus wild-type, Mann-Whitney test). (C) FGFR4 RMS772 mutants showed 3-fold higher invasion (normalized to the vector control) by a modified Boyden chamber assay (t test with Welch correction). (D) Left: Representative IVVM images show the presence of RMS cells in the lung (1 and 24 hour time points shown). Right: Quantification of pulmonary lesions at 1 and 24 hours showed the persistence of RMS cells transduced with the 2 mutants (normalized to the vector control at each time point; n = 5 mice per group; Mann-Whitney test). (E) Gross pulmonary metastases from RMS772 cells at 21 days (comparison using Student’s t test). (F) Left: Kaplan-Meier survival analysis demonstrated a significantly poorer survival in mice injected intravenously with RMS772 cell lines transduced with the 2 mutants. Right: Pulmonary lesions in representative animals injected with mutants are shown.
Figure 8. FGFR4 mutations transform 3T3 cells.
(A) FGFR4 mutants transformed NIH 3T3 cells and promoted in vivo tumor growth after subcutaneous injection in nude mice (*P < 0.001 versus wild type, Mann-Whitney test). Representative animals injected with 3T3 cells transduced with the respective constructs are shown at day 18. In these animals, tumors were observable in 4 of 5 mice (K535) and 5 of 5 mice (E550). (B) Kaplan-Meier survival analysis in mice after intravenous injection of NIH 3T3 cells with wild-type and mutant FGFR4 (n = 5 mice per group).
Figure 9. Oncogene dependence and inhibition of FGFR4.
(A) Cell cycle analysis of RMS772 in 10% serum. (B) Cell cycle analysis in the absence of serum showed increased apoptotic cells (subG1 fraction) for the vector and wild-type FGFR4 RMS772 cell lines. (C) Inhibition of FGFR4 phosphorylation after PD173074 treatment at 3 hours. (D) RMS772 survival after 48 hour treatment with PD173074. IC50 values for RMS772 vector control, wild-type FGFR4, K535, and E550 are presented. (E) Increased subG1 fraction of mutant RMS772 cells lines after 24 hours of treatment with 20 μM PD173074. (F) Increased caspase-3 activation of FGFR4 mutant RMS772 cell lines after 24 hours of treatment with 20 μM PD173074.
Similar articles
- Targeting wild-type and mutationally activated FGFR4 in rhabdomyosarcoma with the inhibitor ponatinib (AP24534).
Li SQ, Cheuk AT, Shern JF, Song YK, Hurd L, Liao H, Wei JS, Khan J. Li SQ, et al. PLoS One. 2013 Oct 4;8(10):e76551. doi: 10.1371/journal.pone.0076551. eCollection 2013. PLoS One. 2013. PMID: 24124571 Free PMC article. - Functional screening of FGFR4-driven tumorigenesis identifies PI3K/mTOR inhibition as a therapeutic strategy in rhabdomyosarcoma.
McKinnon T, Venier R, Yohe M, Sindiri S, Gryder BE, Shern JF, Kabaroff L, Dickson B, Schleicher K, Chouinard-Pelletier G, Menezes S, Gupta A, Zhang X, Guha R, Ferrer M, Thomas CJ, Wei Y, Davani D, Guidos CJ, Khan J, Gladdy RA. McKinnon T, et al. Oncogene. 2018 May;37(20):2630-2644. doi: 10.1038/s41388-017-0122-y. Epub 2018 Feb 28. Oncogene. 2018. PMID: 29487419 Free PMC article. - FGFR4 blockade exerts distinct antitumorigenic effects in human embryonal versus alveolar rhabdomyosarcoma.
Crose LE, Etheridge KT, Chen C, Belyea B, Talbot LJ, Bentley RC, Linardic CM. Crose LE, et al. Clin Cancer Res. 2012 Jul 15;18(14):3780-90. doi: 10.1158/1078-0432.CCR-10-3063. Epub 2012 May 30. Clin Cancer Res. 2012. PMID: 22648271 Free PMC article. - Fibroblast Growth Factor Receptor 4 (FGFR4) Selective Inhibitors as Hepatocellular Carcinoma Therapy: Advances and Prospects.
Lu X, Chen H, Patterson AV, Smaill JB, Ding K. Lu X, et al. J Med Chem. 2019 Mar 28;62(6):2905-2915. doi: 10.1021/acs.jmedchem.8b01531. Epub 2018 Nov 16. J Med Chem. 2019. PMID: 30403487 Review. - Is fibroblast growth factor receptor 4 a suitable target of cancer therapy?
Heinzle C, Erdem Z, Paur J, Grasl-Kraupp B, Holzmann K, Grusch M, Berger W, Marian B. Heinzle C, et al. Curr Pharm Des. 2014;20(17):2881-98. doi: 10.2174/13816128113199990594. Curr Pharm Des. 2014. PMID: 23944363 Free PMC article. Review.
Cited by
- PROX1 transcription factor controls rhabdomyosarcoma growth, stemness, myogenic properties and therapeutic targets.
Gizaw NY, Kallio P, Punger T, Gucciardo E, Haglund C, Böhling T, Lehti K, Sampo M, Alitalo K, Kivelä R. Gizaw NY, et al. Proc Natl Acad Sci U S A. 2022 Dec 6;119(49):e2116220119. doi: 10.1073/pnas.2116220119. Epub 2022 Dec 2. Proc Natl Acad Sci U S A. 2022. PMID: 36459642 Free PMC article. - Targeting the Fibroblast Growth Factor Receptor (FGFR) Family in Lung Cancer.
Pacini L, Jenks AD, Lima NC, Huang PH. Pacini L, et al. Cells. 2021 May 10;10(5):1154. doi: 10.3390/cells10051154. Cells. 2021. PMID: 34068816 Free PMC article. Review. - FGFR-targeted therapeutics: clinical activity, mechanisms of resistance and new directions.
Katoh M, Loriot Y, Brandi G, Tavolari S, Wainberg ZA, Katoh M. Katoh M, et al. Nat Rev Clin Oncol. 2024 Apr;21(4):312-329. doi: 10.1038/s41571-024-00869-z. Epub 2024 Feb 29. Nat Rev Clin Oncol. 2024. PMID: 38424198 Review. - DFG-out mode of inhibition by an irreversible type-1 inhibitor capable of overcoming gate-keeper mutations in FGF receptors.
Huang Z, Tan L, Wang H, Liu Y, Blais S, Deng J, Neubert TA, Gray NS, Li X, Mohammadi M. Huang Z, et al. ACS Chem Biol. 2015 Jan 16;10(1):299-309. doi: 10.1021/cb500674s. Epub 2014 Oct 27. ACS Chem Biol. 2015. PMID: 25317566 Free PMC article. - Artificial neural network inference (ANNI): a study on gene-gene interaction for biomarkers in childhood sarcomas.
Tong DL, Boocock DJ, Dhondalay GK, Lemetre C, Ball GR. Tong DL, et al. PLoS One. 2014 Jul 15;9(7):e102483. doi: 10.1371/journal.pone.0102483. eCollection 2014. PLoS One. 2014. PMID: 25025207 Free PMC article.
References
- Parham D.M., Ellison D.A. Rhabdomyosarcomas in adults and children: an update. Arch. Pathol. Lab. Med. 2006;130:1454–1465. - PubMed
- Raney R.B., et al. Rhabdomyosarcoma and undifferentiated sarcoma in the first two decades of life: a selective review of intergroup rhabdomyosarcoma study group experience and rationale for Intergroup Rhabdomyosarcoma Study V. J. Pediatr. Hematol. Oncol. 2001;23:215–220. doi: 10.1097/00043426-200105000-00008. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous