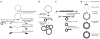Telomeric circles: universal players in telomere maintenance? - PubMed (original) (raw)
. 2009 Oct;16(10):1010-5.
doi: 10.1038/nsmb.1660. Epub 2009 Oct 6.
Affiliations
- PMID: 19809492
- PMCID: PMC4041010
- DOI: 10.1038/nsmb.1660
Telomeric circles: universal players in telomere maintenance?
Lubomir Tomaska et al. Nat Struct Mol Biol. 2009 Oct.
Abstract
To maintain linear DNA genomes, organisms have evolved numerous means of solving problems associated with DNA ends (telomeres), including telomere-associated retrotransposons, palindromes, hairpins, covalently bound proteins and the addition of arrays of simple DNA repeats. Telomeric arrays can be maintained through various mechanisms such as telomerase activity or recombination. The recombination-dependent maintenance pathways may include telomeric loops (t-loops) and telomeric circles (t-circles). The potential involvement of t-circles in telomere maintenance was first proposed for linear mitochondrial genomes. The occurrence of t-circles in a wide range of organisms, spanning yeasts, plants and animals, suggests the involvement of t-circles in many phenomena including the alternative-lengthening of telomeres (ALT) pathway and telomere rapid deletion (TRD). In this Perspective, we summarize these findings and discuss how t-circles may be related to t-loops and how t-circles may have initiated the evolution of telomeres.
Figures
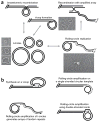
Figure 1
t-circles and t-loops seem to be common players in telomere maintenance. (a) Proposed pathway in the maintenance of mitochondrial telomeres in the yeast C. parapsilosis. The mitochondrial telomeres consist of t-arrays (n × 738 bp) with a 5′ single-stranded overhang of about 110 nt. Invasion of the overhang into the double-stranded region of t-arrays can form t-loop structures. The t-circles can be formed either by intramolecular recombination between repeats within t-arrays or by excision from t-loops. The t-circles amplify autonomously via a rolling-circle mechanism (forming σ-form ‘tailed circles’), thus generating long extrachromosomal t-arrays, which can recombine with the main genome and extend the termini. EM revealed the key players in the proposed pathway of the mitochondrial telomere maintenance,,,,. (b) Proposed role for the rolling-circle synthesis in nuclear telomere maintenance. Rolling-circle synthesis can extend the termini in situ (on a t-loop or single-stranded or double-stranded t-circles), or extrachromosomal t-circles may replicate autonomously.
Figure 2
Evolutionary scenario for the emergence of linear chromosomes terminating with t-arrays and hypotheses on the emergence of t-circles. (a) Genome rearrangements within a circular genome may result in formation of a palindrome. Alternatively, a selfish genetic element may ‘infect’ a circular genome. Subsequent resolution of the palindrome eventually generates a linear molecule with t-hairpins at the ends. The termini represent a substrate for recombination with t-circles and/or elongation by telomerase. Expanded t-arrays allow the formation of higher-order structures such as t-loops and the emergence of telomere maintenance mechanism(s). We have proposed three mechanisms that may explain the emergence of t-circles. (b) Similarities between mitochondrial t-circles and hypersuppresive (ρ −) petite genomes indicate that they could have emerged by a common mechanism. Hypersuppressive genomes represent short fragments of a mitochondrial genome, which replicates via rolling-circle synthesis. In baker’s yeast, hypersuppressive genomes can outcompete the main genome. However, in strictly aerobic _petite_-negative organisms, the two replicons (mtDNA and hypersuppressive-like t-circles) can coexist, and eventually the t-circles can recombine with mtDNA, which may result in the formation of a linear DNA genome with t-arrays at the ends. Rolling-circle replicating t-circles may escape from the mitochondria to the nucleus and analogously affect the nuclear genome. (c) Alternatively, t-circles may emerge from intra-strand recombination of units within a preexisting intrachromosomal array of tandem repeats. (d) T-circles may also emerge by reverse transcription of an RNA molecule (or an RNA fragment, shown in gray), which serves as a template for the first-strand cDNA synthesis, followed by template degradation, synthesis of the second cDNA strand and ligation of nicks. The double-stranded DNA circle is either discarded (most cases) because of its inability to replicate, or if it is able to replicate it can be preserved and may invade into the main genome or chromosome, as shown in a.
Similar articles
- Amplification of telomeric arrays via rolling-circle mechanism.
Nosek J, Rycovska A, Makhov AM, Griffith JD, Tomaska L. Nosek J, et al. J Biol Chem. 2005 Mar 18;280(11):10840-5. doi: 10.1074/jbc.M409295200. Epub 2005 Jan 18. J Biol Chem. 2005. PMID: 15657051 - Recombination at long mutant telomeres produces tiny single- and double-stranded telomeric circles.
Groff-Vindman C, Cesare AJ, Natarajan S, Griffith JD, McEachern MJ. Groff-Vindman C, et al. Mol Cell Biol. 2005 Jun;25(11):4406-12. doi: 10.1128/MCB.25.11.4406-4412.2005. Mol Cell Biol. 2005. PMID: 15899847 Free PMC article. - Alternatives to telomerase: keeping linear chromosomes via telomeric circles.
Tomaska L, McEachern MJ, Nosek J. Tomaska L, et al. FEBS Lett. 2004 Jun 1;567(1):142-6. doi: 10.1016/j.febslet.2004.04.058. FEBS Lett. 2004. PMID: 15165907 Review. - Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain.
Cesare AJ, Groff-Vindman C, Compton SA, McEachern MJ, Griffith JD. Cesare AJ, et al. Mol Cell Biol. 2008 Jan;28(1):20-9. doi: 10.1128/MCB.01122-07. Epub 2007 Oct 29. Mol Cell Biol. 2008. PMID: 17967889 Free PMC article. - Telomeres in evolution and evolution of telomeres.
Fajkus J, Sýkorová E, Leitch AR. Fajkus J, et al. Chromosome Res. 2005;13(5):469-79. doi: 10.1007/s10577-005-0997-2. Chromosome Res. 2005. PMID: 16132812 Review.
Cited by
- Telomere- and Telomerase-Associated Proteins and Their Functions in the Plant Cell.
Procházková Schrumpfová P, Schořová Š, Fajkus J. Procházková Schrumpfová P, et al. Front Plant Sci. 2016 Jun 28;7:851. doi: 10.3389/fpls.2016.00851. eCollection 2016. Front Plant Sci. 2016. PMID: 27446102 Free PMC article. Review. - Sequence characterization of eccDNA content in glyphosate sensitive and resistant Palmer amaranth from geographically distant populations.
Spier Camposano H, Molin WT, Saski CA. Spier Camposano H, et al. PLoS One. 2022 Sep 14;17(9):e0260906. doi: 10.1371/journal.pone.0260906. eCollection 2022. PLoS One. 2022. PMID: 36103503 Free PMC article. - The mitochondrial genome of the pathogenic yeast Candida subhashii: GC-rich linear DNA with a protein covalently attached to the 5' termini.
Fricova D, Valach M, Farkas Z, Pfeiffer I, Kucsera J, Tomaska L, Nosek J. Fricova D, et al. Microbiology (Reading). 2010 Jul;156(Pt 7):2153-2163. doi: 10.1099/mic.0.038646-0. Epub 2010 Apr 15. Microbiology (Reading). 2010. PMID: 20395267 Free PMC article. - Break-Induced Replication: The Where, The Why, and The How.
Kramara J, Osia B, Malkova A. Kramara J, et al. Trends Genet. 2018 Jul;34(7):518-531. doi: 10.1016/j.tig.2018.04.002. Epub 2018 May 4. Trends Genet. 2018. PMID: 29735283 Free PMC article. Review. - Signalling inhibition by ponatinib disrupts productive alternative lengthening of telomeres (ALT).
Kusuma FK, Prabhu A, Tieo G, Ahmed SM, Dakle P, Yong WK, Pathak E, Madan V, Jiang YY, Tam WL, Kappei D, Dröge P, Koeffler HP, Jeitany M. Kusuma FK, et al. Nat Commun. 2023 Apr 6;14(1):1919. doi: 10.1038/s41467-023-37633-3. Nat Commun. 2023. PMID: 37024489 Free PMC article.
References
- Gilson E, Géli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. - PubMed
- McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu Rev Genet. 2000;34:331–358. - PubMed
- Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. - PubMed
- Nosek J, Kosa P, Tomaska L. On the origin of telomeres: a glimpse at the pre-telomerase world. Bioessays. 2006;28:182–190. - PubMed
- Pardue ML, DeBaryshe PG. Drosophila telomeres: a variation on the telomerase theme. In: Nosek J, Tomaska L, editors. Origin and Evolution of Telomeres. Landes Bioscience; Austin, Texas: 2008. pp. 27–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 55005622/HHMI/Howard Hughes Medical Institute/United States
- R01 ES013773/ES/NIEHS NIH HHS/United States
- R03 TW005654/TW/FIC NIH HHS/United States
- R01 GM031819/GM/NIGMS NIH HHS/United States
- 2-R03-TW005654-04A1/TW/FIC NIH HHS/United States
- ES13773/ES/NIEHS NIH HHS/United States
- P01 CA019014/CA/NCI NIH HHS/United States
- GM31819/GM/NIGMS NIH HHS/United States
- R56 ES013773/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases