The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions - PubMed (original) (raw)
Comparative Study
. 2009 Nov 1;183(9):5896-908.
doi: 10.4049/jimmunol.0803285. Epub 2009 Oct 7.
Affiliations
- PMID: 19812203
- PMCID: PMC2819326
- DOI: 10.4049/jimmunol.0803285
Comparative Study
The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions
Con Sullivan et al. J Immunol. 2009.
Abstract
Mammalian immune responses to LPS exposure are typified by the robust induction of NF-kappaB and IFN-beta responses largely mediated by TLR4 signal transduction pathways. In contrast to mammals, Tlr4 signal transduction pathways in nontetrapods are not well understood. Comprehensive syntenic and phylogenetic analyses support our hypothesis that zebrafish tlr4a and tlr4b genes are paralogous rather than orthologous to human TLR4. Furthermore, we provide evidence to support our assertion that the in vivo responsiveness of zebrafish to LPS exposure is not mediated by Tlr4a and Tlr4b paralogs because they fail to respond to LPS stimulation in vitro. Zebrafish Tlr4a and Tlr4b paralogs were also unresponsive to heat-killed Escherichia coli and Legionella pneumophila. Using chimeric molecules in which portions of the zebrafish Tlr4 proteins were fused to portions of the mouse TLR4 protein, we show that the lack of responsiveness to LPS was most likely due to the inability of the extracellular portions of zebrafish Tlr4a and Tlr4b to recognize the molecule, rather than to changes in their capacities to transduce signals through their Toll/IL-1 receptor (TIR) domains. Taken together, these findings strongly support the notion that zebrafish tlr4a and tlr4b paralogs have evolved to provide alternative ligand specificities to the Tlr immune defense system in this species. These data demonstrate that intensive examination of gene histories when describing the Tlr proteins of basally diverging vertebrates is required to obtain fuller appreciation of the evolution of their function. These studies provide the first evidence for the functional evolution of a novel Tlr.
Conflict of interest statement
Disclosures: The authors have no financial conflict of interest.
Figures
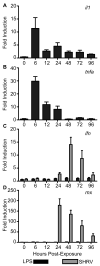
Figure 1. Expression of immune-responsive genes upon exposure to known TLR4 agonist LPS
Zebrafish aged 20-30 dpf were exposed to 10 μg·mL-1 LPS for 24 h or 106TCID50 ·mL-1 SHRV for 5 h. qRT-PCR was performed on first-strand cDNA reverse transcribed from total RNA collected from zebrafish that had been sacrificed at the indicated times. Changes in gene expression, measured as fold induction ± SEM, were recorded for (A) il1, (B) tnfa, (C) ifn, and (D) mx at 0, 6, 12, 24, 48, 72, and 96 hpe. Elevated levels of transcription were observed for il1 (A) and tnfa (B) upon LPS exposure, but not for ifn (C) and mx (D).
Figure 2. Amino acid alignment of human, mouse and zebrafish TLR4 homologs
Amino acid sequences for human TLR4 (Accession No. U93091 [
http://www.ncbi.nlm.nih.gov/nuccore/2429102
]), mouse TLR4 (Accession No. Q9QUK6 [
http://www.ncbi.nlm.nih.gov/protein/20140894
]), zebrafish Tlr4a (Accession No. ACE74929 [
http://www.ncbi.nlm.nih.gov/protein/190341705
]), and zebrafish Tlr4b (Accession No. AAH68358 [
http://www.ncbi.nlm.nih.gov/protein/46249703
]) were aligned using AlignX, a ClustalW-based algorithm that is part of the VectorNTI software package. Dashes indicate gaps in aligned sequence. The red overlines indicate the LRR domain; the blue overline indicates the predicted transmembrane domain; and the purple overlines identify the TIR domain. The color scheme of the alignment is the default output of the AlignX software: black on white indicates a non-similar residue for which no consensus sequence could be determined; blue on cyan denotes a consensus residue from a block of similar residues at the aligned position; black on green indicates a consensus residue with equal to or greater than 50% identity at the aligned position; red on yellow denotes a consensus residue with 100% identity to the other residues at the aligned position; and green on white describes a residue with weak similarity to the consensus residue at a given position.
Figure 3. Zebrafish Tlr4 proteins form a clade with human, mouse, and Gallus TLR4s
A broad representation of TLR proteins were aligned with AlignX. Extracellular domains were extracted and subjected to molecular phylogenetic analysis using PHYLIP, software version 3.6b. The TLR4 proteins from human, mouse, Gallus, and zebrafish form a monophyletic group with 100% bootstrap support.
Figure 4. Conserved syntenies surrounding zebrafish tlr4 genes
(A) Human chromosome 10, showing three regions that contain orthologs of the indicated tlr4 neighbors of zebrafish tlr4 genes. In each of the three regions, the indicated genes are either adjacent to each other or are within two genes of each other. (B) A 0.5 megabase region of zebrafish LG13 containing tlr4a and tlr4b. Labels such as 558624 PDZD7 10q24.3 indicate the annotated designation of the zebrafish sequence, the abbreviation of the human ortholog, and the location of the human ortholog. Results show that the _tlr4_-containing portion of the zebrafish genome is orthologous to parts of human chromosome 10. (C) 50kb of pufferfish chromosome 17, demonstrating co-linear orthologies with the corresponding segment of the zebrafish genome, with an exception for the absence of the two tlr4 genes. The most parsimonious explanation is that the tlr4 gene region was deleted from the pufferfish lineage after the divergence of the zebrafish and pufferfish lineages.
Figure 5. Paralogons for the zebrafish chromosome segment containing tlr4 genes
(A) Paralogies for human chromosome 10, which is pictured at the top with the locations of several orthologs of genes neighboring zebrafish tlr4 genes (orthologs to STOX1, CCAR1, and CXXC6 [left red rectangle], PDZD7 and SFXN3 [middle red rectangle], and C10ORF58 and MAT1A [right red rectangles]). Yellow bars represent regions of Hsa2, 4, 5, and 8 paralogous to the corresponding portion of Hsa10q. (B) Hsa9, the location of TLR4. (C) A 3.3 Mb portion of Hsa9q22 containing TLR4 and the closest human paralog to PDZD7. (D) A 0.5 Mb portion of zebrafish LG13 containing the tlr4 genes. (E) A 2.2 Mb segment of Hsa4q35 containing STOX2, the closest paralog to stox1, and TLR3. (F) EIF4E, the human ortholog of the zebrafish tlr4 neighbor zgc:110154, resides on Hsa4q with TLR2 and TLR3. These data would be expected according to the hypothesis that the zebrafish tlr4 genes are paralogs, not orthologs, of the human TLR4 gene.
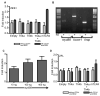
Figure 6. Zebrafish Tlr4a and Tlr4b are unresponsive to LPS stimulation
Zebrafish Tlr4a and Tlr4b, alone and in combination, were overexpressed in 293H (A) cells, in the presence or absence of human CD14 and human MD2. mTLR4 was similarly overexpressed to serve as a positive control. Each was expressed in the presence or absence of human CD14 and MD2 in order to facilitate the mTLR4 positive and negative control, respectively. In the absence of MD2 and CD14, pcDNA3.1 empty vector was overexpressed to maintain an equal amount of plasmid in the transfection. pBIIx-luc (to measure NF-κB reporter activity) and pRL-CMV (to normalize luciferase activity data) were also co-transfected. Following transfection, cells were exposed to ultrapure LPS (10 μg · mL-1) for 6 h and then lysed. Firefly and Renilla relative luminescence units were recorded. The data are presented as the normalized mean fold induction of NF-κB-luciferase reporter activity ± SEM. (B) RT-PCR reveals expression of myd88, ticam1 (trif), and tirap (mal) in ZFL cells. Total RNA was extracted from ZFL cells and reverse transcribed to cDNA (+). A mock reverse transcription, in which the reverse transcriptase was excluded, was also performed (-). PCR was performed using gene-specific primers for myd88, ticam1 (trif), and tirap (mal) (see supplementary Table S1C). (C) Full-length zebrafish myd88 was subcloned in frm2bl so that the egfp was replaced. ZFL cells were transfected with 10, 100, and 400 ng of myd88 plasmid, along with empty frm (no egfp) so that the total frm plasmid transfected was 400 ng. In addition, 400 ng of pBIIx-luc and 10 ng of pRL-CMV were co-transfected. Forty-eight hours post-transfection, cells were lysed, and luciferase activities were measured. The data are presented as normalized mean fold induction of NF-κB-luciferase reporter activity ± SEM over empty vector control. (D) As in (A), zebrafish Tlr4a and Tlr4b, alone and in combination, were overexpressed in ZFL cells, in the presence or absence of human CD14 and human MD2. mTLR4 was overexpressed as a positive control. pBIIx-luc and pRL-CMV were also co-transfected. Following transfection, cells were exposed to ultrapure LPS (10 μg · mL-1) for 6 h and then lysed. Firefly and Renilla relative luminescence units were recorded. The data are presented as the normalized mean fold induction of NF-κB-luciferase reporter activity ± SEM.
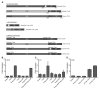
Figure 7. The lack of LPS responsiveness is due to the extracellular domains of Tlr4a and Tlr4b
(A) Schematic of native and chimeric Tlr4 constructs created by PCR sewing of DNA sequence encoding the mTLR4 extracellular domain to the transmembrane and intracellular domains of zebrafish Tlr4a and Tlr4b or the zebrafish Tlr4a and Tlr4b extracellular domains to the transmembrane and intracellular domains of mTLR4. The extracellular domain, including the LRR region, the transmembrane (TM) domain, and the intracellular domains, including the Toll-IL1 receptor (TIR) domain, for the native and chimeric proteins are labeled accordingly. Numbers below each of the native proteins indicate the N-terminus of the protein (left), the N-terminal portion of the transmembrane domain (middle), and the C-termimus of the protein (right). (B,C) Plasmids encoding Tlr4aΔLRR and Tlr4bΔLRR proteins were overexpressed in 293H (B) and ZFL (C), and the relative induction of an NF-κB-reporter was measured relative to an empty vector control. Native Tlr4a and Tlr4b proteins were overexpressed for comparison. To determine if Tlr4aΔLRR and Tlr4bΔLRR synergize, each was overexpressed in a manner such that the total amount of plasmid transfected did not exceed that which was transfected for the individual constructs (i.e. 200 ng of each, 400 ng total). (D) Chimeric Tlr4 constructs were overexpressed in 293H cells, along with CD14, MD2, pBIIx-luc, and pRL-CMV. Cells were exposed to LPS (10 μg · mL-1) for 6 h and then lysed. Firefly and Renilla relative luminescence units were recorded. The data are presented as the normalized mean fold induction of NF-κB-luciferase reporter activity ± SEM.
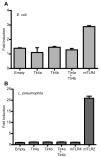
Figure 8. Zebrafish Tlr4a and Tlr4b are unresponsive to stimulation by heat-killed E. coli or L. pneumophila
Constructs encoding zebrafish Tlr4a and Tlr4b, alone and in combination, were overexpressed in 293H cells along with CD14, MD2, pBIIx-luc, and pRL-CMV. For HKEC exposure, empty vector was transfected as a negative control, and plasmid encoding mTLR4 was transfected as a positive control. For HKLP exposure, empty vector and plasmid encoding mTLR4 were transfected as negative controls, and plasmid encoding mTLR2 was transfected as a positive control. Following transfection, cells were exposed to (A) HKEC (106 CFU · mL-1) or (B) HKLP (108 CFU · mL-1) for 6 h and then lysed. Firefly and Renilla relative luminescence units were recorded. The data are presented as the normalized mean fold induction of NF-κB luciferase reporter activity ± SEM.
Figure 9. A model for the historical relationships of human and zebrafish _TLR4_-related genes
Zebrafish tlr4 genes arose from a common ancestral TLR4 gene. Following two rounds of genome duplication, TLR4A and TLR4B appeared. With lineage divergence, there was reciprocal loss of _TLR4 ohno_logs, leading to the appearance of the TLR4A gene now present in the mammalian genome (known as TLR4) and tlr4b in the zebrafish/pufferfish ancestor. Following an additional _ohno_log loss, the tlr4b gene disappeared from the pufferfish lineage. In contrast, the tlr4b gene was duplicated in the zebrafish lineage, leading to the appearance of tlr4ba (currently known as tlr4a) and tlr4bb (currently known as tlr4b).
References
- Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. - PubMed
- Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. - PubMed
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. - PubMed
- Poltorak A, Smirnova I, He X, Liu MY, Van Huffel C, McNally O, Birdwell D, Alejos E, Silva M, Du X, Thompson P, Chan EK, Ledesma J, Roe B, Clifton S, Vogel SN, Beutler B. Genetic and physical mapping of the Lps locus: identification of the toll-4 receptor as a candidate gene in the critical region. Blood Cells Mol Dis. 1998;24:340–355. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R15 AI049237/AI/NIAID NIH HHS/United States
- R15AI065509-01/AI/NIAID NIH HHS/United States
- R15AI049237-02/AI/NIAID NIH HHS/United States
- R01 GM087308/GM/NIGMS NIH HHS/United States
- R01 RR10715/RR/NCRR NIH HHS/United States
- R15 AI065509/AI/NIAID NIH HHS/United States
- R01 RR010715/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases