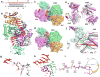Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes - PubMed (original) (raw)
Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes
Yanli Wang et al. Nature. 2009.
Abstract
The slicer activity of the RNA-induced silencing complex resides within its Argonaute (Ago) component, in which the PIWI domain provides the catalytic residues governing guide-strand mediated site-specific cleavage of target RNA. Here we report on structures of ternary complexes of Thermus thermophilus Ago catalytic mutants with 5'-phosphorylated 21-nucleotide guide DNA and complementary target RNAs of 12, 15 and 19 nucleotides in length, which define the molecular basis for Mg(2+)-facilitated site-specific cleavage of the target. We observe pivot-like domain movements within the Ago scaffold on proceeding from nucleation to propagation steps of guide-target duplex formation, with duplex zippering beyond one turn of the helix requiring the release of the 3'-end of the guide from the PAZ pocket. Cleavage assays on targets of various lengths supported this model, and sugar-phosphate-backbone-modified target strands showed the importance of structural and catalytic divalent metal ions observed in the crystal structures.
Conflict of interest statement
Conflict of interest
TT is a cofounder and scientific advisor to Alnylam Pharmaceuticals and an advisor to Regulus Therapeutics.
Figures
Figure 1. Crystal structure of T. thermophilus Ago containing N546 catalytic mutant bound to 5′-phosphorylated 21-nt guide DNA and 12-nt target RNA
a, Sequence of the guide DNA-target RNA duplex. The traceable segments of the bases of the guide DNA and target RNA in the structure of the ternary complex are shown in red and blue, respectively. Disordered segments of the bases on both strands that cannot be traced are shown in grey. b, A view of the 2.6 Å crystal structure of the Ago ternary complex. The Ago protein is color-coded by domains (N in cyan, PAZ in magenta, Mid in orange and PIWI in green) and linkers (L1 and L2 in grey). The bound 21-nt guide DNA is in red and traced for bases 1–12 and 20–21, whereas the bound 12-nt target RNA is in blue and traced for bases 2′–12′. Backbone phosphorus atoms are in yellow. Both ends of the bound guide DNA are anchored in this ternary complex. c, An expanded view of the ternary complex highlighting the alignment of guide DNA (1–3) and target RNA (2′–3′), where the bases of the 1–2 step of the guide strand are splayed apart. Note the intermolecular hydrogen-bonding of the Watson-crick edge of T1 with the backbone amide carbonyl of M413 and side chain of N436, as well as the positioning of phosphate 1 of the guide strand in the Mid binding pocket. A Mg2+ cation (purple) coordinates to phosphates 1 and 3 of the guide strand, as well as to an inserted carboxylate of V685 from the C-terminus of the protein. d, An expanded view of the ternary complex highlighting the guide DNA (1–12) - target RNA (2′–12′) duplex, together with the catalytic residues (D478, D660 and N546 mutant) of the RNase H fold of the PIWI domain. The scissile phosphate group at the 10–11 step of the target RNA is indicated by a red arrow. e, An expanded view highlighting the positioning of the backbone phosphate linking the 10–11 step (phosphorus colored in magenta) of the target RNA relative to the catalytic residues (D478, D660 and N546 mutant) in the ternary complex. f, Positioning of the side chain of R548 relative to the guide DNA (6 to 12) - target RNA (6′ to 12′) duplex. Note the intermolecular contacts between the sugar-phosphate backbone of the guide strand and side chains of the protein in the ternary complex.
Figure 2. Crystal structure of T. thermophilus Ago containing E546 catalytic mutant bound to 5′-phosphorylated 21-nt guide DNA and 15-nt target RNA
a, Sequence of the guide DNA-target RNA duplex, with traceable segments color-coded as in Fig. 1a. b, A view of the 3.05 Å crystal structure of the Ago ternary complex, color-coded as outlined in Fig. 1b. The bound 21-nt guide DNA is in red and traced for bases 1–16, whereas the bound 15-nt target RNA is in blue and traced for bases 2′–15′. Only the 5′-end of the guide DNA is anchored in this ternary complex. Comparison of crystal structures of c, Ago N546 mutant - 12-mer target and d, Ago E546 mutant - 15-nt target ternary complexes. The Ago protein is shown in a surface representation with labeled domains and linkers color-coded as in Fig. 1b. The guide DNA (red) and target RNA (blue) are shown in stick representation with backbone phosphorus atoms in yellow. e, View of alignment of Ago N546 mutant - 12-mer target complex (magenta) and Ago E546 mutant - 15-nt target complex (silver), after superpositioning of their PIWI-containing (Mid and PIWI domains) modules. The yellow arrow indicates the magnitude of the conformational change on proceeding from the 12-mer target to 15-mer target ternary complexes. f, Conformational changes in loop 1 (residue 479-488, red arrow) and loop 2 (residues 505-516, green arrow) of the PIWI domain on proceeding from the 12-nt target ternary complex (magenta) to the 15-nt target ternary complex (silver). Only the DNA-RNA duplex for the 15-nt target ternary complex is shown in cyan in a surface representation. Loops 1 and 2 are colored in light red (labeled L1′) and light green (labeled L2′) in the 12-nt target ternary complex, while they are colored in dark red (labeled L1) and dark green (labeled L2) in the 15-nt target ternary complex. The β-strand involved in sliding is highlighted by black double-edged arrow. g, The ternary complex containing 12-nt target RNA. Residues 11 and 12 of the guide strand are in red and loops L1′ and L2′ are in light red and light green, respectively. h, The ternary complex containing 15-nt target RNA. Residues 11 and 15 of the guide strand are in red and loops L1 and L2 are in dark red and dark green, respectively. Loop L1 switches to a β-turn aligned through hydrogen bonding both within the turn and also with loop L2, thereby stabilizing this turn conformation. The main-chain of Glu512 forms a hydrogen bond with phosphate group of residue 14 of the guide DNA. The positively-charged side chains of Arg513 and Arg486 interact with the backbone of the DNA guide strand, as indicated by blue arrows. i, A ribbon representation of the sliding of the β-strand (Gly489 to Val494) by one residue and conformational transition in adjacent L1 loop on proceeding from the 12-mer RNA target ternary complex (magenta) to 15-mer target RNA ternary complex (silver).
Figure 3. Crystal structure of T. thermophilus Ago containing N478 catalytic mutant bound to 5′-phosphorylated 21-nt guide DNA and 19-nt target RNA and identification of Mg2+ binding sites within the catalytic pocket of the wild-type Ago complex
a, Sequence of the guide DNA-target RNA duplex, with traceable segments color-coded as in Fig. 1a. b, A view of the 2.8 Å crystal structure of the ternary complex, color-coded as outlined in Fig. 1b. The bound 21-nt guide DNA is in red and traced for bases 1–16, whereas the bound 19-nt RNA is in blue and traced for bases 2′–16′. Only the 5′-end of the guide strand is anchored in this ternary complex. c, An expanded view of the 19-mer target ternary complex highlighting blocking of propagation of the guide DNA - target RNA duplex beyond pair 16 by the N domain. Base 16 of the guide strand stacks over the aromatic ring of Tyr43, while base 16′ stacks over Pro44. d, Intermolecular hydrogen-bonding contacts between the sugar-phosphate backbone of the 10′ to 13′ target RNA segment and backbone and side chains of the PIWI domain in the 19-mer target ternary complex. e, f, Fo-Fc omit maps (blue color, contoured at 3.5σ) of the 9′–12′ segment of bound RNA and catalytic D478, D546 and D660 residues in the 3.3 Å structures of the ternary complexes in 50 mM Mg2+ (space group P43212, one molecule in the asymmetric unit, panel e) and in 80 mM Mg2+ (space group P212121, two molecules in asymmetric unit, panel f). Bound Mg2+ cation(s) were identified in omit maps contoured in purple at 6.0σ as outlined in (e) and (f), based on coordination to several oxygen atoms in an approximate octahedral geometry. One bound Mg2+ cation can be assigned in the ternary complex in 50 mM Mg2+ in (e) and two bound Mg2+ cations can be assigned in the ternary complex in 80 mM Mg2+ in (f).
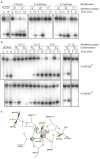
Figure 4. Effect of complementarity and length on target DNA cleavage by T. thermophilus Ago
0.5 μM 5′-phosphorylated guide oligodeoxynucleotides were incubated with 0.5 μM T. thermophilus Ago at 55 °C for 30 min in the presence of 5 mM Mg2+ followed by the addition of 0.5 μM 5′-32P-radiolabeled (*) DNA target and further incubation at 75 °C for the indicated time. Cleavage products were resolved on denaturing polyacrylamide gels and visualised by phosphoimaging; for DNA sequences, see Supplementary Table 4. a, Schematic of the DNA duplex that was manipulated in this experiment; the cleavage site is indicated by an arrow, the position of the 32P label by an asterix. b, Shortening of the target DNA from its 5′ end. Alterations of the target DNA and corresponding paired structure are illustrated to the left. Target DNA cleavage was performed at 65 °C rather than 75 °C to facilitate hybridization of shortened targets. c, Positional variation of 15-nt target DNAs. For labelling and reaction conditions, see panel b.
Figure 5. Effect of sugar-phosphate backbone modifications on target DNA cleavage by T. thermophilus Ago
Cleavage experiments were performed as described in Fig. 4. a, 2′-Fluoro-, 2′-methoxy-, and 2′-hydroxyl-substitutions of single 2′-deoxyribose residues of the target DNA strand at and nearby the cleavage site. The control target (unmod.) was the unmodified oligodeoxynucleotide. b, Phosphorothioate modification of the target DNA. The phosphate configuration (RP or SP) of the phosphorothioate diastereomers is indicated. Cleavage assays were performed in the presence of either Mg2+ or Mn2+ cations. Note that the experiment for the 11′–12′ isomers was a different experiment, where overall reaction rates were slower. For the complete experiment see Supplementary Fig. 25. For sequences of oligonucleotides see Supplementary Table 4. c, Structure of the cleavage site modelling the attack of the hydroxyl nucleophile. Phosphate oxygen and active site carboxylate oxygens coordinated to metal ions A and B (purple spheres), with distances less than 2.5 Å are shown as blue dashed lines. The coordination of the carboxylate oxygen from D546 to metal ion B is hidden in the projection. The phosphate oxygens and 2′ residues sensitive to modification are shown in yellow and green spheres, respectively; R = 2′-H, OH, F, or Ome. The red arrows indicate the attack of the hydroxyl nucleophile modelled to be directly coordinated by metal ion A, and the stabilization of the developing negative charge of the 3′ oxyanion leaving group by metal ion B.
Comment in
- Structural biology: Tracing Argonaute binding.
Bouasker S, Simard MJ. Bouasker S, et al. Nature. 2009 Oct 8;461(7265):743-4. doi: 10.1038/461743a. Nature. 2009. PMID: 19812664 No abstract available.
Similar articles
- Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex.
Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Wang Y, et al. Nature. 2008 Dec 18;456(7224):921-6. doi: 10.1038/nature07666. Nature. 2008. PMID: 19092929 Free PMC article. - Structural biology: Tracing Argonaute binding.
Bouasker S, Simard MJ. Bouasker S, et al. Nature. 2009 Oct 8;461(7265):743-4. doi: 10.1038/461743a. Nature. 2009. PMID: 19812664 No abstract available. - Structure of the guide-strand-containing argonaute silencing complex.
Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Wang Y, et al. Nature. 2008 Nov 13;456(7219):209-13. doi: 10.1038/nature07315. Epub 2008 Aug 27. Nature. 2008. PMID: 18754009 Free PMC article. - When Argonaute takes out the ribonuclease sword.
Nakanishi K. Nakanishi K. J Biol Chem. 2024 Jan;300(1):105499. doi: 10.1016/j.jbc.2023.105499. Epub 2023 Nov 27. J Biol Chem. 2024. PMID: 38029964 Free PMC article. Review. - Molecular mechanism of target RNA transcript recognition by Argonaute-guide complexes.
Parker JS, Roe SM, Barford D. Parker JS, et al. Cold Spring Harb Symp Quant Biol. 2006;71:45-50. doi: 10.1101/sqb.2006.71.029. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381279 Review.
Cited by
- DNA-guided genome editing using the Natronobacterium gregoryi Argonaute.
Gao F, Shen XZ, Jiang F, Wu Y, Han C. Gao F, et al. Nat Biotechnol. 2016 Jul;34(7):768-73. doi: 10.1038/nbt.3547. Epub 2016 May 2. Nat Biotechnol. 2016. PMID: 27136078 Retracted. - Structural and mechanistic insights into a mesophilic prokaryotic Argonaute.
Tao X, Ding H, Wu S, Wang F, Xu H, Li J, Zhai C, Li S, Chen K, Wu S, Liu Y, Ma L. Tao X, et al. Nucleic Acids Res. 2024 Oct 28;52(19):11895-11910. doi: 10.1093/nar/gkae820. Nucleic Acids Res. 2024. PMID: 39315697 Free PMC article. - A Dynamic Search Process Underlies MicroRNA Targeting.
Chandradoss SD, Schirle NT, Szczepaniak M, MacRae IJ, Joo C. Chandradoss SD, et al. Cell. 2015 Jul 2;162(1):96-107. doi: 10.1016/j.cell.2015.06.032. Cell. 2015. PMID: 26140593 Free PMC article. - Large domain motions in Ago protein controlled by the guide DNA-strand seed region determine the Ago-DNA-mRNA complex recognition process.
Xia Z, Huynh T, Ren P, Zhou R. Xia Z, et al. PLoS One. 2013;8(1):e54620. doi: 10.1371/journal.pone.0054620. Epub 2013 Jan 29. PLoS One. 2013. PMID: 23382927 Free PMC article. - Advances in RNA Interference for Plant Functional Genomics: Unveiling Traits, Mechanisms, and Future Directions.
Chaudhary D, Jeena AS, Rohit, Gaur S, Raj R, Mishra S, Kajal, Gupta OP, Meena MR. Chaudhary D, et al. Appl Biochem Biotechnol. 2024 Sep;196(9):5681-5710. doi: 10.1007/s12010-023-04850-x. Epub 2024 Jan 4. Appl Biochem Biotechnol. 2024. PMID: 38175411 Review.
References
- Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457–467. - PubMed
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. - PubMed
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. - PubMed
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 19:517–529. - PubMed
- Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM068476-05/GM/NIGMS NIH HHS/United States
- R01 AI068776-04/AI/NIAID NIH HHS/United States
- R01 AI068776-05/AI/NIAID NIH HHS/United States
- P30 EB009998/EB/NIBIB NIH HHS/United States
- R01 GM068476/GM/NIGMS NIH HHS/United States
- R01 AI068776/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources