Evolution of CLE signaling: origins of the CLV1 and SOL2/CRN receptor diversity - PubMed (original) (raw)
Evolution of CLE signaling: origins of the CLV1 and SOL2/CRN receptor diversity
Hiroki Miwa et al. Plant Signal Behav. 2009 Jun.
Abstract
The shoot apical meristem is maintained by the intercellular factor, CLV3, a dodecapeptide in Arabidopsis. CLV3 belongs to the CLE family and putative CLE genes have been found in various plants, even in the moss Physcomitrella patens. Here, we report that a pteridophyte, Selaginella moelendorffii, also has 15 putative CLE genes in its genome. On the other hand, CLV1 is reported to function as a receptor for the CLV3 peptide, and other CLE peptides might be recognized by CLV1 homologues in various plants. Recent genetic studies of the crn and sol2 mutants of Arabidopsis have revealed that SOL2/CRN encodes a receptor-like kinase protein. SOL2/CRN functions together with CLV2 independently of CLV1 in the CLE signaling pathway. Phylogenetic analysis of CLV1, CLV2 and SOL2/CRN revealed that Arabidopsis, rice, Populus trichocarpa and Vitis vinifera have one copy of the SOL2/CRN and CLV2 homologues, and Selaginella moelendorffii and Physcomitrella patens have no homologues. In contrast, a number of CLV1 homologues were identified in the genomic databases of Arabidopsis, rice, Populus trichocarpa, Vitis vinifera, and even a pteridophyte, Selaginella moelendorffii, and a moss, Physcomitrella patens. These results indicate that CLV1 and its homologues play multiple roles in plant development and environmental responses, whereas SOL2/CRN and CLV2 have more specific roles in vascular plants.
Figures
Figure 1
Alignment of the deduced polypeptides of the Selaginella CLE gene family and a Physcomitrella PpCLE1 gene. The conserved dodecapeptide CLE region is shown in boldface. The sequences of CLE genes are available in the DDBJ database (
http://www.ddbj.nig.ac.jp/index-e.html
). Accession numbers: SmCLE1, AB465350; SmCLE2, AB465351; SmCLE3, AB465352; SmCLE4, AB465353; SmCLE5, AB465354; SmCLE6, AB465355; SmCLE7, AB465356; SmCLE8, AB465357; SmCLE9, AB465358; SmCLE10, AB465359; SmCLE11, AB465360; SmCLE12, AB465361; SmCLE13, AB465362; SmCLE14, AB465363; SmCLE15, AB465364.
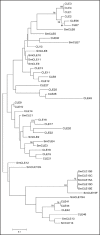
Figure 2
An unrooted neighbor-joining tree of CLE dodecapeptides. Twelve-amino-acid sequences of the CLE peptide regions (Fig. 1) encoded by 15 Selaginella genes and 31 Arabidopsis CLE genes were used to construct the phylogenic tree. Bootstrap values of 30% and above, from the neighbor-joining method with Kimura's correction, are shown. The scale bar indicates the number of amino acid substitutions per site.
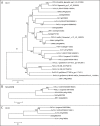
Figure 3
Comparison of SOL2/CRN, CLV2, CLV1, and their homologues. Phylogenetic relationships of SOL2/CRN, CLV2 and CLV1 to their counterparts from other plant species, Arabidopsis, rice, Populus trichocarpa, Vitis vinifera, Selaginella moelendorffii and Physcomitrella patens. The tree was calculated and drawn with the MEGA 3.1 program from an alignment of complete protein sequences, with gap deletion. Bootstrap values of 50% and above, from the neighbor-joining method with Kimura's correction, are shown. The amino-acid sequences of AtBRI1 (A and C) and AtCLV1 (B) were used as an outgroup. The scale bar indicates the number of amino acid substitutions per site. The cut-off of E-value for SOL2/CRN and CLV2 homologs is e−100. The cut-off of E-value for Selaginella CLV1 homologs is e−184.
Similar articles
- CLE peptide signaling during plant development.
Wang G, Fiers M. Wang G, et al. Protoplasma. 2010 Apr;240(1-4):33-43. doi: 10.1007/s00709-009-0095-y. Epub 2009 Dec 17. Protoplasma. 2010. PMID: 20016993 Free PMC article. Review. - RPK2 functions in diverged CLE signaling.
Sawa S, Tabata R. Sawa S, et al. Plant Signal Behav. 2011 Jan;6(1):86-8. doi: 10.4161/psb.6.1.14128. Epub 2011 Jan 1. Plant Signal Behav. 2011. PMID: 21301217 Free PMC article. - The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis.
Miwa H, Betsuyaku S, Iwamoto K, Kinoshita A, Fukuda H, Sawa S. Miwa H, et al. Plant Cell Physiol. 2008 Nov;49(11):1752-7. doi: 10.1093/pcp/pcn148. Epub 2008 Oct 14. Plant Cell Physiol. 2008. PMID: 18854335 Free PMC article. - CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification.
Guo Y, Han L, Hymes M, Denver R, Clark SE. Guo Y, et al. Plant J. 2010 Sep;63(6):889-900. doi: 10.1111/j.1365-313X.2010.04295.x. Plant J. 2010. PMID: 20626648 Free PMC article. - CLAVATA3, a plant peptide controlling stem cell fate in the meristem.
Hirakawa Y. Hirakawa Y. Peptides. 2021 Aug;142:170579. doi: 10.1016/j.peptides.2021.170579. Epub 2021 May 24. Peptides. 2021. PMID: 34033873 Review.
Cited by
- CLE peptide signaling during plant development.
Wang G, Fiers M. Wang G, et al. Protoplasma. 2010 Apr;240(1-4):33-43. doi: 10.1007/s00709-009-0095-y. Epub 2009 Dec 17. Protoplasma. 2010. PMID: 20016993 Free PMC article. Review. - The Plant Peptidome: An Expanding Repertoire of Structural Features and Biological Functions.
Tavormina P, De Coninck B, Nikonorova N, De Smet I, Cammue BP. Tavormina P, et al. Plant Cell. 2015 Aug;27(8):2095-118. doi: 10.1105/tpc.15.00440. Epub 2015 Aug 14. Plant Cell. 2015. PMID: 26276833 Free PMC article. Review. - Peptide signaling in plant development.
Katsir L, Davies KA, Bergmann DC, Laux T. Katsir L, et al. Curr Biol. 2011 May 10;21(9):R356-64. doi: 10.1016/j.cub.2011.03.012. Curr Biol. 2011. PMID: 21549958 Free PMC article. Review. - The Function of the CLE Peptides in Plant Development and Plant-Microbe Interactions.
Betsuyaku S, Sawa S, Yamada M. Betsuyaku S, et al. Arabidopsis Book. 2011;9:e0149. doi: 10.1199/tab.0149. Epub 2011 Sep 26. Arabidopsis Book. 2011. PMID: 22303273 Free PMC article. - RPK2 functions in diverged CLE signaling.
Sawa S, Tabata R. Sawa S, et al. Plant Signal Behav. 2011 Jan;6(1):86-8. doi: 10.4161/psb.6.1.14128. Epub 2011 Jan 1. Plant Signal Behav. 2011. PMID: 21301217 Free PMC article.
References
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. - PubMed
- Kinoshita A, Nakamura Y, Sasaki E, Kyozuka J, Fukuda H, Sawa S. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007;48:1821–1825. - PubMed
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, et al. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous