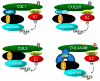CRL4s: the CUL4-RING E3 ubiquitin ligases - PubMed (original) (raw)
Review
CRL4s: the CUL4-RING E3 ubiquitin ligases
Sarah Jackson et al. Trends Biochem Sci. 2009 Nov.
Abstract
The evolutionarily conserved cullin family proteins can assemble as many as 400 distinct E3 ubiquitin ligase complexes that regulate diverse cellular pathways. CUL4, one of three founding cullins conserved from yeast to humans, uses a large beta-propeller protein, DDB1, as a linker to interact with a subset of WD40 proteins that serve as substrate receptors, forming as many as 90 E3 complexes in mammals. Many CRL4 complexes are involved in chromatin regulation and are frequently hijacked by different viruses.
Figures
Figure 1. Assembly of Cullin–RING ubiquitin ligases (CRLs)
The cullin-ROC family of E3 ligases regulates the ubiquitylation of many substrates by assembling into multiple distinct E3 ligases. Each cullin uses a modular assembly to recruit different substrates to a common catalytic core by varying its substrate receptor. Cullin family members (green) from different organisms share similar mechanisms for assembling a multi-subunit complex to ubiquitylate specific substrate protein (light blue). An N-terminal domain interacts either directly with a protein motif (orange) present in substrate receptors (black) or via a linker (blue). Separately, a C-terminal domain binds with a small RING protein (ROC1 or ROC2, yellow) which recruits and allosterically activates an E2 enzyme (red) that transfers ubiquitin (Ub; color) to the substrate. Cullins are activated by the covalent conjugation with a ubiquitin-like modifier, NEDD8 (Nd8; purple). Human cells express an estimated 78 F-box proteins, 205 BTB proteins, about 90 DWD proteins and an estimated 50 BC-box proteins.
Figure 2. Schematic comparison of CUL4 proteins and mouse Cul4A mutants
A. Human cells contain two CUL4 genes encoding CUL4A (NP_001008895) and two isoforms of CUL4B (NP_001073341 and NP_003579.3) proteins which differ by only 22 amino acids at the N-terminus. B. Wild type mouse Cul4A gene and comparison of three targeted Cul4A mutants. Filled and open boxes denote coding and non-coding exons, respectively. Exons encoding the DDB1 and ROC 1 binding regions in CUL4A have been noted. Three mouse models targeting Cul4A have been generated to date, deleting exon 1, exons 4-8, and exons 17-19, respectively. Discrepancies in reported phenotypes might be due to unintentional disruption of a nearby neighboring gene, Pcid2.
Similar articles
- Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery.
Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Angers S, et al. Nature. 2006 Oct 5;443(7111):590-3. doi: 10.1038/nature05175. Nature. 2006. PMID: 16964240 - DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases.
He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. He YJ, et al. Genes Dev. 2006 Nov 1;20(21):2949-54. doi: 10.1101/gad.1483206. Genes Dev. 2006. PMID: 17079684 Free PMC article. - Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes.
Zhang Y, Feng S, Chen F, Chen H, Wang J, McCall C, Xiong Y, Deng XW. Zhang Y, et al. Plant Cell. 2008 Jun;20(6):1437-55. doi: 10.1105/tpc.108.058891. Epub 2008 Jun 13. Plant Cell. 2008. PMID: 18552200 Free PMC article. - Ubiquitin proteasome system (UPS): what can chromatin do for you?
O'Connell BC, Harper JW. O'Connell BC, et al. Curr Opin Cell Biol. 2007 Apr;19(2):206-14. doi: 10.1016/j.ceb.2007.02.014. Epub 2007 Feb 20. Curr Opin Cell Biol. 2007. PMID: 17314036 Review. - DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase.
Lee J, Zhou P. Lee J, et al. Mol Cell. 2007 Jun 22;26(6):775-80. doi: 10.1016/j.molcel.2007.06.001. Mol Cell. 2007. PMID: 17588513 Review.
Cited by
- Artemis interacts with the Cul4A-DDB1DDB2 ubiquitin E3 ligase and regulates degradation of the CDK inhibitor p27.
Yan Y, Zhang X, Legerski RJ. Yan Y, et al. Cell Cycle. 2011 Dec 1;10(23):4098-109. doi: 10.4161/cc.10.23.18227. Epub 2011 Dec 1. Cell Cycle. 2011. PMID: 22134138 Free PMC article. - A novel function of CRL4(Cdt2): regulation of the subunit structure of DNA polymerase δ in response to DNA damage and during the S phase.
Zhang S, Zhao H, Darzynkiewicz Z, Zhou P, Zhang Z, Lee EY, Lee MY. Zhang S, et al. J Biol Chem. 2013 Oct 11;288(41):29550-61. doi: 10.1074/jbc.M113.490466. Epub 2013 Aug 2. J Biol Chem. 2013. PMID: 23913683 Free PMC article. - Lentivirus Vpr and Vpx accessory proteins usurp the cullin4-DDB1 (DCAF1) E3 ubiquitin ligase.
Romani B, Cohen EA. Romani B, et al. Curr Opin Virol. 2012 Dec;2(6):755-63. doi: 10.1016/j.coviro.2012.09.010. Epub 2012 Oct 10. Curr Opin Virol. 2012. PMID: 23062609 Free PMC article. Review. - Cullin-4B E3 ubiquitin ligase mediates Apaf-1 ubiquitination to regulate caspase-9 activity.
Ohta E, Itoh M, Ueda M, Hida Y, Wang MX, Hayakawa-Ogura M, Li S, Nishida E, Ohta K, Tana, Islam S, Nakagawa K, Sunayama T, Chen H, Hirata S, Endo M, Ohno Y, Nakagawa T. Ohta E, et al. PLoS One. 2019 Jul 22;14(7):e0219782. doi: 10.1371/journal.pone.0219782. eCollection 2019. PLoS One. 2019. PMID: 31329620 Free PMC article. - CRL4Mahj E3 ubiquitin ligase promotes neural stem cell reactivation.
Ly PT, Tan YS, Koe CT, Zhang Y, Xie G, Endow S, Deng WM, Yu F, Wang H. Ly PT, et al. PLoS Biol. 2019 Jun 6;17(6):e3000276. doi: 10.1371/journal.pbio.3000276. eCollection 2019 Jun. PLoS Biol. 2019. PMID: 31170139 Free PMC article.
References
- Pickart CM. Mechanisms Underlying Ubiquitination. Annu Rev Biochem. 2001;70:503–533. - PubMed
- Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. - PubMed
- Willems AR, et al. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. - PubMed
- Kipreos ET, et al. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous