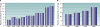Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms - PubMed (original) (raw)
Review
Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms
Philip D Lister et al. Clin Microbiol Rev. 2009 Oct.
Abstract
Treatment of infectious diseases becomes more challenging with each passing year. This is especially true for infections caused by the opportunistic pathogen Pseudomonas aeruginosa, with its ability to rapidly develop resistance to multiple classes of antibiotics. Although the import of resistance mechanisms on mobile genetic elements is always a concern, the most difficult challenge we face with P. aeruginosa is its ability to rapidly develop resistance during the course of treating an infection. The chromosomally encoded AmpC cephalosporinase, the outer membrane porin OprD, and the multidrug efflux pumps are particularly relevant to this therapeutic challenge. The discussion presented in this review highlights the clinical significance of these chromosomally encoded resistance mechanisms, as well as the complex mechanisms/pathways by which P. aeruginosa regulates their expression. Although a great deal of knowledge has been gained toward understanding the regulation of AmpC, OprD, and efflux pumps in P. aeruginosa, it is clear that we have much to learn about how this resourceful pathogen coregulates different resistance mechanisms to overcome the antibacterial challenges it faces.
Figures
FIG. 1.
Increasing prevalence of multidrug resistance among P. aeruginosa isolates from ICU patients in the United States. (A) Data for 13,999 nonduplicate isolates collected from 1993 to 2002 (178); (B) data for 37,390 isolates collected from 1997 to 2000 (132). Data represent the percentage of P. aeruginosa isolates that expressed a phenotype of multidrug resistance (resistance to three or more drug classes) during each year of the studies. (Panel A is adapted from reference with permission; panel B is based on data from reference .)
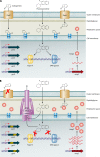
FIG. 2.
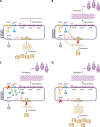
Mutational resistance to fluoroquinolones and carbapenems involving chromosomally encoded mechanisms expressed by P. aeruginosa. (A) Interactions of fluoroquinolones and carbapenems with “wild-type” susceptible P. aeruginosa expressing basal levels of AmpC, OprD, and nonmutated fluoroquinolone target genes (gyrA, gyrB, parC, and parE). Fluoroquinolone molecules pass through the outer membrane, peptidoglycan, periplasmic space, and cytoplasmic membrane and interact with DNA gyrase and topoisomerase IV (Topo IV) targets in the cytoplasm when these enzymes are complexed with DNA. Carbapenem molecules pass through the outer membrane-specific porin OprD and interact with their target PBPs, located on the outside of the cytoplasmic membrane. (B) Chromosomally encoded mechanisms of resistance to fluoroquinolones and carbapenems. Fluoroquinolone resistance is mediated by (i) overexpression of RND efflux pumps extruding the drug molecules from the periplasmic and cytoplasmic spaces and/or (ii) mutational changes within the target genes. Locations of the QRDRs within target genes are highlighted in yellow. Carbapenem resistance is mediated primarily by (i) decreased production or loss of functional OprD in the outer membrane and/or (ii) overproduction of RND efflux pumps (with the exception of imipenem). Minor changes in susceptibility can be observed due to overexpression of AmpC, adding to the resistance potential.
FIG. 3.
Mechanisms involved in regulation of ampC expression. These figures represent the current knowledge obtained from studies with members of the Enterobacteriaceae and appear to parallel events in P. aeruginosa. (A) Wild-type basal expression of ampC. During normal cell wall recycling, 1,6-anhydromuropeptides are removed from the cell wall and transported into the cytoplasm via the AmpG permease. The 1,6-anhydromuropeptides are cleaved by AmpD to generate free tripeptides, which are later converted into UDP-MurNAc-pentapeptides. UDP-MurNAc-pentapeptide interacts with AmpR bound to the _ampR_-ampC intergenic region, creating a conformation that represses transcription of ampC. Low basal levels of AmpC are produced, and the enzyme is localized to the periplasmic space. (B) β-Lactam induction of ampC expression. Inducing β-lactams, such as cefoxitin and imipenem, cross the outer membrane through porins, enter the periplasmic space, and interact with target PBPs. An increase in pools of 1,6-anhydromuropeptides is observed, and AmpD is unable to efficiently process the higher levels of cell wall fragments. The anhydro-MurNAc-peptides (inducing peptides) replace UDP-MurNAc-pentapeptides (suppressing peptides) bound to AmpR, causing a conformational change in the protein. AmpR is converted into a transcriptional activator, ampC is expressed at higher levels, and levels of AmpC increase in the periplasmic space. When the amount of β-lactam decreases below “inducing levels,” the cytoplasmic pool of anhydro-MurNAc-peptides also decreases, and AmpD is able to efficiently cleave these peptides, restoring wild-type ampC expression, as shown in panel A. (C) AmpD-associated derepression of ampC expression. Mutations leading to the inactivation of AmpD or decreased expression of ampD impair the processing of cell wall recycled products and lead to increased levels of anhydro-MurNAc-peptides (inducing peptides) in the cytoplasm. As a result, the binding of inducing peptides to AmpR is favored, AmpR is “locked” in a conformation for transcriptional activation of ampC expression, and high-level constitutive expression of ampC is observed.
FIG. 3.
Mechanisms involved in regulation of ampC expression. These figures represent the current knowledge obtained from studies with members of the Enterobacteriaceae and appear to parallel events in P. aeruginosa. (A) Wild-type basal expression of ampC. During normal cell wall recycling, 1,6-anhydromuropeptides are removed from the cell wall and transported into the cytoplasm via the AmpG permease. The 1,6-anhydromuropeptides are cleaved by AmpD to generate free tripeptides, which are later converted into UDP-MurNAc-pentapeptides. UDP-MurNAc-pentapeptide interacts with AmpR bound to the _ampR_-ampC intergenic region, creating a conformation that represses transcription of ampC. Low basal levels of AmpC are produced, and the enzyme is localized to the periplasmic space. (B) β-Lactam induction of ampC expression. Inducing β-lactams, such as cefoxitin and imipenem, cross the outer membrane through porins, enter the periplasmic space, and interact with target PBPs. An increase in pools of 1,6-anhydromuropeptides is observed, and AmpD is unable to efficiently process the higher levels of cell wall fragments. The anhydro-MurNAc-peptides (inducing peptides) replace UDP-MurNAc-pentapeptides (suppressing peptides) bound to AmpR, causing a conformational change in the protein. AmpR is converted into a transcriptional activator, ampC is expressed at higher levels, and levels of AmpC increase in the periplasmic space. When the amount of β-lactam decreases below “inducing levels,” the cytoplasmic pool of anhydro-MurNAc-peptides also decreases, and AmpD is able to efficiently cleave these peptides, restoring wild-type ampC expression, as shown in panel A. (C) AmpD-associated derepression of ampC expression. Mutations leading to the inactivation of AmpD or decreased expression of ampD impair the processing of cell wall recycled products and lead to increased levels of anhydro-MurNAc-peptides (inducing peptides) in the cytoplasm. As a result, the binding of inducing peptides to AmpR is favored, AmpR is “locked” in a conformation for transcriptional activation of ampC expression, and high-level constitutive expression of ampC is observed.
FIG. 4.
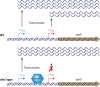
Characterization of oprD promoter elements. Transcription of oprD in P. aeruginosa PAO1 initiates with equal frequencies from two start sites, located 23 bases (SS1) and 71 bases (SS2) upstream of the structural gene (ATG translation start codon is highlighted in orange). The putative −10 and −35 promoter elements for SS1 are highlighted in red, and the putative −10 and −35 promoter elements for SS2 are highlighted in blue.
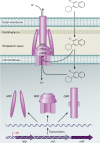
FIG. 5.
Structure and function of RND efflux pumps in P. aeruginosa. RND pumps typically exist in a tripartite system consisting of an RND cytoplasmic membrane transporter (RND), an MFP, and an OMF. This complex forms a channel spanning the entire membrane, allowing for the proton-driven transport of lipophilic and amphiphilic drugs from the cytoplasm of the cell across the cytoplasmic membrane, through the periplasmic space, across the peptidoglycan, and across the outer membrane. The RND efflux pumps can also extrude drugs from the periplasmic space before they cross the cytoplasmic membrane.
FIG. 6.
RND efflux operons in P. aeruginosa. Genes which encode protein components or characterized pumps are denoted by their gene names, and genes encoding protein components of uncharacterized pumps are designated with the P. aeruginosa (PA) numbers assigned in the annotated P. aeruginosa genome sequence (GenBank). Genes are depicted with the following color scheme: dark red arrow, transcriptional regulator-encoding gene; dark blue arrow, membrane fusion protein-encoding gene; light blue arrow, RND transporter-encoding gene; red arrow, outer membrane protein-encoding gene; and gold arrow, gene encoding a protein with unknown function. (Adapted from reference with permission of the publisher.)
FIG. 7.
Coregulation of mexEF-oprN and oprD in P. aeruginosa. The models represent the proposed mechanisms of coregulation of mexEF-oprN and oprD in P. aeruginosa. Each panel represents the chromosome of P. aeruginosa, highlighting the mexE, mexF, oprN, and oprD structural genes and the proposed genes involved in coregulation, mexT, mexS, and mvaT. (A) Basal expression of mexEF-oprN and oprD in wild-type P. aeruginosa. In wild-type P. aeruginosa, MexT is functionally inactive due to either the presence of suppressing mutations or the lack of a secondary effector molecule. As a result, expression of _mexEF_-oprN occurs at a low basal level, and expression of oprD occurs at a level sufficient to provide quantities of OprD in the outer membrane sufficient for normal cellular function. (B) MexT-associated coregulation of mexEF-oprN and oprD. In _nfxC_-type mutants, MexT becomes active through a mutation within the structural gene. The activated MexT protein positively regulates (green arrow) transcription of _mexEF_-oprN, leading to overexpression of the efflux operon and overproduction of the MexEF-OprN efflux pump. Simultaneously, MexT negatively regulates (red arrow) oprD at the transcriptional and posttranscriptional levels, leading to decreased production of OprD. (C) MexS-associated coregulation of mexEF-oprN and oprD. Loss of MexS, a putative oxidoreductase/dehydrogenase, has been suggested to cause a buildup of secondary metabolites which may serve as effector molecules for MexT. These effector molecules could bind to MexT, alter the conformational state of the regulatory protein, and transform MexT into an activating transcriptional regulator. As a result, MexT can positively regulate (green arrow) the expression of mexEF-oprN and negatively regulate (red arrow) the expression of oprD, similar to what is described for panel B. (D) MvaT-associated coregulation of mexEF-oprN and oprD. Loss of the global regulatory protein MvaT is also associated with the upregulation of the _mexEF_-oprN operon. The mechanism of MvaT-associated regulation has not been elucidated, but it functions independent of MexT and MexS. In contrast to the case for the MexT- and MexS-associated regulatory pathways, loss of MvaT causes an upregulation of both mexEF-oprN and oprD expression.
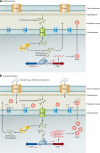
FIG. 8.
MexT-associated downregulation of oprD expression. Transcription of oprD in wild-type P. aeruginosa PAO1 initiates from two start sites, SS1 and SS2. MexT-associated downregulation of oprD expression is associated with a selective inhibition of transcription from SS1 (278). The model proposed in this figure involves the binding of a regulatory protein, potentially MexT, with or without cofactors, within the promoter region between SS1 and SS2, blocking efficient initiation of transcription from SS1.
Similar articles
- Chromosomally-encoded resistance mechanisms of Pseudomonas aeruginosa: therapeutic implications.
Lister PD. Lister PD. Am J Pharmacogenomics. 2002;2(4):235-43. doi: 10.2165/00129785-200202040-00003. Am J Pharmacogenomics. 2002. PMID: 12421094 Review. - Role of efflux pumps: MexAB-OprM and MexXY(-OprA), AmpC cephalosporinase and OprD porin in non-metallo-β-lactamase producing Pseudomonas aeruginosa isolated from cystic fibrosis and burn patients.
Aghazadeh M, Hojabri Z, Mahdian R, Nahaei MR, Rahmati M, Hojabri T, Pirzadeh T, Pajand O. Aghazadeh M, et al. Infect Genet Evol. 2014 Jun;24:187-92. doi: 10.1016/j.meegid.2014.03.018. Epub 2014 Mar 30. Infect Genet Evol. 2014. PMID: 24694825 - OprD mutations and inactivation, expression of efflux pumps and AmpC, and metallo-β-lactamases in carbapenem-resistant Pseudomonas aeruginosa isolates from South Korea.
Lee JY, Ko KS. Lee JY, et al. Int J Antimicrob Agents. 2012 Aug;40(2):168-72. doi: 10.1016/j.ijantimicag.2012.04.004. Epub 2012 May 26. Int J Antimicrob Agents. 2012. PMID: 22633564 - Phenotypic and genetic resistance traits of Pseudomonas aeruginosa strains infecting cystic fibrosis patients: A French cohort study.
Courtois N, Caspar Y, Maurin M. Courtois N, et al. Int J Antimicrob Agents. 2018 Sep;52(3):358-364. doi: 10.1016/j.ijantimicag.2018.05.008. Epub 2018 Jul 18. Int J Antimicrob Agents. 2018. PMID: 29775685 - Mechanisms of β-lactam resistance among Pseudomonas aeruginosa.
Wolter DJ, Lister PD. Wolter DJ, et al. Curr Pharm Des. 2013;19(2):209-22. Curr Pharm Des. 2013. PMID: 22894618 Review.
Cited by
- Targeted Genome Reduction of Pseudomonas aeruginosa Strain PAO1 Led to the Development of Hypovirulent and Hypersusceptible rDNA Hosts.
Grosjean M, Guénard S, Giraud C, Muller C, Plésiat P, Juarez P. Grosjean M, et al. Front Bioeng Biotechnol. 2021 Mar 11;9:640450. doi: 10.3389/fbioe.2021.640450. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 33777913 Free PMC article. - Could Azithromycin Be Part of Pseudomonas aeruginosa Acute Pneumonia Treatment?
Leroy AG, Caillon J, Caroff N, Broquet A, Corvec S, Asehnoune K, Roquilly A, Crémet L. Leroy AG, et al. Front Microbiol. 2021 Mar 16;12:642541. doi: 10.3389/fmicb.2021.642541. eCollection 2021. Front Microbiol. 2021. PMID: 33796090 Free PMC article. Review. - Ceftazidime-avibactam activity against multidrug-resistant Pseudomonas aeruginosa isolated in U.S. medical centers in 2012 and 2013.
Sader HS, Castanheira M, Mendes RE, Flamm RK, Farrell DJ, Jones RN. Sader HS, et al. Antimicrob Agents Chemother. 2015;59(6):3656-9. doi: 10.1128/AAC.05024-14. Epub 2015 Apr 6. Antimicrob Agents Chemother. 2015. PMID: 25845861 Free PMC article. - Quorum-Sensing Mechanisms and Bacterial Response to Antibiotics in P. aeruginosa.
Rasamiravaka T, El Jaziri M. Rasamiravaka T, et al. Curr Microbiol. 2016 Nov;73(5):747-753. doi: 10.1007/s00284-016-1101-1. Epub 2016 Jul 23. Curr Microbiol. 2016. PMID: 27449213 Review. - Eugenol-Functionalized Magnetite Nanoparticles Modulate Virulence and Persistence in Pseudomonas aeruginosa Clinical Strains.
Mohammed HB, Rayyif SMI, Curutiu C, Birca AC, Oprea OC, Grumezescu AM, Ditu LM, Gheorghe I, Chifiriuc MC, Mihaescu G, Holban AM. Mohammed HB, et al. Molecules. 2021 Apr 10;26(8):2189. doi: 10.3390/molecules26082189. Molecules. 2021. PMID: 33920270 Free PMC article.
References
- Abdalhamid, B., P. A. Wickman, and N. D. Hanson. 2005. Correlation of ampC induction with PBP binding in Enterobacter cloacae, abstr. C1-2211. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother., Washington, DC.
- Aendekerk, S., S. P. Diggle, Z. Song, N. Hoiby, P. Cornelis, P. Williams, and M. Camara. 2005. The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology 151:1113-1125. - PubMed
- Aendekerk, S., B. Ghysels, P. Cornelis, and C. Baysse. 2002. Characterization of a new efflux pump, MexGHI-OpmD from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 148:2371-2381. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources