Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization - PubMed (original) (raw)
Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization
Saurabh Sen et al. J Biol Chem. 2009.
Abstract
Dominant missense mutations in the leucine-rich repeat kinase 2 (LRRK2) gene are the most common known genetic cause of Parkinson disease. LRRK2 encodes a serine/threonine protein kinase, and pathogenic mutations may increase kinase activity. Intrinsic GTP binding in the GTPase domain may govern kinase activity through an internal signal transduction cascade. As with many protein kinases, LRRK2 self-interacts through mechanisms that may regulate enzymatic activity. We find that the disruption of either GTPase or kinase activity enhances the formation of high molecular weight oligomers and prevents the formation of LRRK2 dimer structures. In addition, brief application of the broad spectrum kinase inhibitor staurosporine ablates LRRK2 dimers and promotes LRRK2 high molecular weight oligomers. LRRK2 interactions with other proteins in cell lines are kinase-independent and include chaperones and cell cytoskeleton components, suggesting that LRRK2 self-assembly principally dictates complex size. To further explore the mechanics of kinase activation, we separate soluble LRRK2 protein that encodes the pathogenic G2019S mutation into high molecular weight oligomers, dimers, and monomers and find that kinase activity resides with dimeric LRRK2. Some PD-associated mutations that increase kinase activity in vitro significantly increase the proportion of dimer structures relative to total LRRK2 protein, providing additional insight into how pathogenic mutations may alter normal enzymatic regulation. Targeting and tracking LRRK2 dimerization may provide a clear way to observe LRRK2 kinase activity in living cells, and disruption of dimeric LRRK2 through kinase inhibition or other means may attenuate pathogenic increases in LRRK2 enzymatic output.
Figures
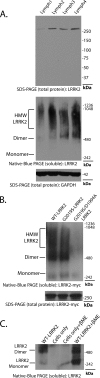
FIGURE 1.
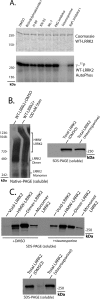
LRRK2 forms kinase-sensitive high molecular weight conformations. A, cell pellets derived from four lymphoblast cell lines (labeled Lymph1 to -4) were split equally for lysis by freeze/thaw cycles directly in PBS or lysis with 1% SDS and PBS with sonication, and 10 μg of protein lysate (as determined by BCA protein assay) was loaded onto a native gel (3–12% bis-tris) or an SDS gel (7% Tris acetate-SDS), respectively. LRRK2 protein or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was visualized after transfer onto PVDF membrane and incubation with an antibody specific for LRRK2 or glyceraldehyde-3-phosphate dehydrogenase. Complexes consistent with the expected size of a LRRK2 homodimer and LRRK2 monomer are indicated. B, HEK-293T cells were transfected with constructs encoding human wild-type LRRK2, G2019S-LRRK2, or LRRK2 containing both the G2019S and D1994A (kinase-dead) mutations. Cell pellets were split equally for lysis by freeze/thaw cycles directly in PBS or lysis with 1% SDS and PBS with sonication, and 10 μg of protein lysate (as determined by BCA protein assay) was loaded onto a native gel (3–12% bis-tris) or an SDS gel (7% Tris acetate-SDS), respectively. LRRK2-positive complexes were visualized after transfer onto PVDF membrane and incubation with anti-Myc-HRP antibody. C, native lysates of LRRK2-transfected HEK-293FT cells derived from freeze/thaw lysis and subsequently treated with 5% β-ME and heat (65 °C for 10 min) as indicated were analyzed by native PAGE, and complexes consistent with the expected sizes of a LRRK2 homodimer and LRRK2 monomer are indicated.
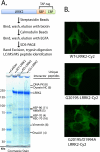
FIGURE 2.
LRRK2 protein interactions and subcellular distribution are independent of LRRK2-kinase activity. A, LRRK2-positive protein complexes were purified from native lysates derived from HEK-293FT cells transfected with WT-LRRK2, kinase-dead (D1994A)-LRRK2, or kinase-overactive G2019S-LRRK2 constructs, through a tandem affinity purification strategy as illustrated. LRRK2 and interacting proteins were resolved from affinity beads, analyzed on a Tris acetate SDS gel, and stained with Coomassie dye, and the constituency of individual protein bands was identified by mass spectrometry. All unique proteins with more than two unique peptide hits of high quality are indicated, and in every case identified proteins matched the expected size in reference to the protein ladder. B, confocal microscopic analysis of HEK-293T cells transfected with the indicated plasmid. Cells were fixed, and LRRK2 expression was detected with anti-Myc-Cy2 antibody (shown as green signal), and cell images are representative. SBP, streptavidin-binding peptide; CBP, calmodulin-binding peptide.
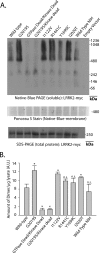
FIGURE 3.
PD-associated LRRK2 mutations enhance the proportion of soluble LRRK2 dimer-sized and high molecular weight species. A, HEK-293T cells were transfected with constructs encoding human LRRK2 that harbors the indicated mutation, where GTPase-dead is K1347A-LRRK2 and kinase-dead is D1994A-LRRK2. A vector encoding WT-LRRK2 (Wild Type-NIH) derived independently from LRRK2-vectors used in this study was provided by Mark Cookson. Cell pellets were split equally for lysis by freeze/thaw cycles directly in PBS or lysis with 1% SDS and PBS with sonication, and 10 μg of protein lysate (as determined by BCA protein assay) was loaded onto a native gel (3–12% bis-tris) or an SDS gel (7% Tris acetate-SDS), respectively. Ponceau S stain was applied to PVDF membranes after transfer of protein complexes to PVDF to ensure even transfer, and the region near 500 kDa is shown and demonstrates the presence of protein across the membrane. LRRK2 complexes were visualized with the anti-c-Myc antibody by Western blot and imaged on a LI-COR Odyssey. Western blots representative of five independent experiments are shown. B, normalization of the LRRK2 dimer-sized structure (signal from ∼480 to ∼550 kDa) to total LRRK2 protein (SDS-solubilized) using densitometry analysis. Error bars, ±S.E. *, p < 0.009. n.s., nonsignificant in comparison with wild-type LRRK2, assessed by unpaired Student's t test.
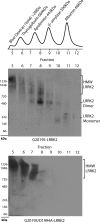
FIGURE 4.
Separation of stable and soluble LRRK2 conformations by size exclusion chromatography. Total soluble native protein lysates derived from cells expressing the indicated protein (kinase-active G2019S-LRRK2 or kinase-inactive G2019S/D1994A-LRRK2) were separated with a Superdex 200 10/300 GL column into seven 1-ml fractions and immediately loaded onto a native gel for analysis by Western blot with the anti-c-Myc-HRP antibody. The _A_280 chromatogram of protein standards (Sigma) previously analyzed through the Superdex column is aligned with fraction number and corresponds to samples loaded onto native gels, whereas native PAGE markers derive from the native mark protein ladder set (Invitrogen). Blue dextran defines the void volume of this column. HMW, high molecular weight.
FIGURE 5.
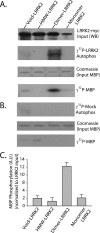
Dimer-sized G2019S-LRRK2 complexes are kinase-active conformations. A, total soluble native protein lysates derived from cells expressing G2019S-LRRK2 were separated with a Superdex 200 10/300 GL column. LRRK2 protein complexes from fractions 5 (void), 8 (dimer), or 10 (monomer) were immunoprecipitated with magnetic beads conjugated to anti-Myc antibody and combined into a kinase reaction containing dephosphorylated MBP protein that serves as an efficient LRRK2 substrate in vitro. Supernatant containing MBP was analyzed by SDS-PAGE and Coomassie stain. Eluted G2019S-LRRK2 protein was resolved onto SDS-gels, transferred to PVDF, and exposed to autoradiography film, and input levels from kinase reactions were determined with anti-Myc-HRP antibody. B, mock kinase reactions using fractionated lysate and immunoprecipitates from cells transected with empty vector and autoradiography films exposed for identical times as in A. C, quantification of LRRK2-mediated MBP phosphorylation via densitometry, adjusted for background and normalized to relative input levels of LRRK2 protein. Data are derived from three independent experiments, and error bars represent ±S.E. A.U., arbitrary units.
FIGURE 6.
Pharmacological inhibition of LRRK2 dimer-sized complexes. A, in vitro measurements of LRRK2 autophosphorylation in the presence of the indicated compound at a concentration of 100 n
m
or DMSO control (0.1% reaction volume). B, HEK-293T cells expressing human WT-LRRK2 protein were treated with the broad spectrum kinase inhibitor staurosporine at a concentration of 100 n
m
, as indicated, for 1 h prior to cell lysis via freeze/thaw cycles in native buffer (PBS); lysate was immediately analyzed by both native PAGE and SDS-PAGE; and LRRK2 expression was visualized by anti-Myc-HRP antibody. Results are representative of four independent experiments. C, native protein lysates prepared as in B were separated by size exclusion chromatography, and fractions enriched in high molecular weight (HMW) oligomer, dimer, or monomer are indicated, and LRRK2 levels in each fraction were determined by SDS-PAGE. Total LRRK2 protein levels in lysates before separation were also verified by SDS-PAGE, with total levels unaffected by staurosporine treatment.
FIGURE 7.
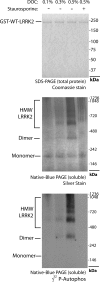
LRRK2 autophosphorylation occurs in dimeric but not monomeric recombinant GST-WT-LRRK2 protein purified from baculovirus-infected insect cells. 200 ng of highly purified and active GST-tagged (N-terminal) truncated WT-LRRK2 protein (obtained from Invitrogen), with purity assessed by SDS-PAGE and Coomassie stain, was combined into a kinase reaction supplemented with varying concentrations of deoxycholate (DOC) that stabilize lower molecular weight LRRK2 oligomers visualized by native PAGE and staurosporine (100 n
m
) that prevents the stabilization of lower molecular weight LRRK2 oligomers but not monomers. Autophosphorylation (i.e. incorporation of 32P) activity is visualized through exposure of dried silver-stained gels to autoradiography film. Kinase activity correlates with dimeric and oligomeric LRRK2 protein but not inactive monomeric LRRK2. HMW, high molecular weight.
FIGURE 8.
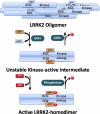
Hypothetical model of LRRK2 kinase activation. A major fraction of LRRK2 protein in cells may reside in large oligomers with low or no kinase activity. LRRK2 oligomers are dissociated through conformational changes induced by GTP binding within the ROC GTPase domain, which may lead to the formation of a dimer structure initially stabilized by a ROC-ROC interaction that unmasks kinase activity and the potential for autophosphorylation. LRRK2 autophosphorylation activity may lead to the stabilization of the kinase-active dimer, which can be destabilized by competing phosphatase activity, GTPase hydrolytic activity, or stochastic interactions with LRRK2 oligomers. In this model, kinase activity is dependent on GTPase activity, whereas GTPase activity is not dependent on kinase activity but must be influenced by changes induced by autophosphorylation. Thus, this model predicts a reciprocal interaction whereby kinase activity and autophosphorylation stabilize lower molecular weight conformations and decrease the reversion to high molecular weight oligomers.
Similar articles
- Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson's disease R1441C/G mutants.
Li X, Tan YC, Poulose S, Olanow CW, Huang XY, Yue Z. Li X, et al. J Neurochem. 2007 Oct;103(1):238-47. doi: 10.1111/j.1471-4159.2007.04743.x. Epub 2007 Jul 10. J Neurochem. 2007. PMID: 17623048 Free PMC article. - GTPase activity regulates kinase activity and cellular phenotypes of Parkinson's disease-associated LRRK2.
Biosa A, Trancikova A, Civiero L, Glauser L, Bubacco L, Greggio E, Moore DJ. Biosa A, et al. Hum Mol Genet. 2013 Mar 15;22(6):1140-56. doi: 10.1093/hmg/dds522. Epub 2012 Dec 13. Hum Mol Genet. 2013. PMID: 23241358 - Kinase activity of mutant LRRK2 manifests differently in hetero-dimeric vs. homo-dimeric complexes.
Leandrou E, Markidi E, Memou A, Melachroinou K, Greggio E, Rideout HJ. Leandrou E, et al. Biochem J. 2019 Feb 8;476(3):559-579. doi: 10.1042/BCJ20180589. Biochem J. 2019. PMID: 30670570 - Contribution of GTPase activity to LRRK2-associated Parkinson disease.
Tsika E, Moore DJ. Tsika E, et al. Small GTPases. 2013 Jul-Sep;4(3):164-70. doi: 10.4161/sgtp.25130. Epub 2013 Jun 10. Small GTPases. 2013. PMID: 24025585 Free PMC article. Review. - Biochemical and molecular features of LRRK2 and its pathophysiological roles in Parkinson's disease.
Seol W. Seol W. BMB Rep. 2010 Apr;43(4):233-44. doi: 10.5483/bmbrep.2010.43.4.233. BMB Rep. 2010. PMID: 20423607 Review.
Cited by
- Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of alpha-synuclein, and apoptotic cell death in aged mice.
Tong Y, Yamaguchi H, Giaime E, Boyle S, Kopan R, Kelleher RJ 3rd, Shen J. Tong Y, et al. Proc Natl Acad Sci U S A. 2010 May 25;107(21):9879-84. doi: 10.1073/pnas.1004676107. Epub 2010 May 10. Proc Natl Acad Sci U S A. 2010. PMID: 20457918 Free PMC article. - LRRK2 inhibition attenuates microglial inflammatory responses.
Moehle MS, Webber PJ, Tse T, Sukar N, Standaert DG, DeSilva TM, Cowell RM, West AB. Moehle MS, et al. J Neurosci. 2012 Feb 1;32(5):1602-11. doi: 10.1523/JNEUROSCI.5601-11.2012. J Neurosci. 2012. PMID: 22302802 Free PMC article. - LRRK2 Biology from structure to dysfunction: research progresses, but the themes remain the same.
Berwick DC, Heaton GR, Azeggagh S, Harvey K. Berwick DC, et al. Mol Neurodegener. 2019 Dec 21;14(1):49. doi: 10.1186/s13024-019-0344-2. Mol Neurodegener. 2019. PMID: 31864390 Free PMC article. Review. - LRRK2 GTPase dysfunction in the pathogenesis of Parkinson's disease.
Xiong Y, Dawson VL, Dawson TM. Xiong Y, et al. Biochem Soc Trans. 2012 Oct;40(5):1074-9. doi: 10.1042/BST20120093. Biochem Soc Trans. 2012. PMID: 22988868 Free PMC article. Review. - VIK-Mediated Auxin Signaling Regulates Lateral Root Development in Arabidopsis.
Shang E, Wei K, Lv B, Zhang X, Lin X, Ding Z, Leng J, Tian H, Ding Z. Shang E, et al. Adv Sci (Weinh). 2024 Sep;11(33):e2402442. doi: 10.1002/advs.202402442. Epub 2024 Jul 3. Adv Sci (Weinh). 2024. PMID: 38958531 Free PMC article.
References
- Cookson M. R. (2005) Annu. Rev. Biochem. 74, 29–52 - PubMed
- Moore D. J., West A. B., Dawson V. L., Dawson T. M. (2005) Annu. Rev. Neurosci. 28, 57–87 - PubMed
- Lesage S., Dürr A., Tazir M., Lohmann E., Leutenegger A. L., Janin S., Pollak P., Brice A. (2006) N. Engl. J. Med. 354, 422–423 - PubMed
- Hulihan M. M., Ishihara-Paul L., Kachergus J., Warren L., Amouri R., Elango R., Prinjha R. K., Upmanyu R., Kefi M., Zouari M., Sassi S. B., Yahmed S. B., El Euch-Fayeche G., Matthews P. M., Middleton L. T., Gibson R. A., Hentati F., Farrer M. J. (2008) Lancet Neurol. 7, 591–594 - PubMed
- Healy D. G., Falchi M., O'Sullivan S. S., Bonifati V., Durr A., Bressman S., Brice A., Aasly J., Zabetian C. P., Goldwurm S., Ferreira J. J., Tolosa E., Kay D. M., Klein C., Williams D. R., Marras C., Lang A. E., Wszolek Z. K., Berciano J., Schapira A. H., Lynch T., Bhatia K. P., Gasser T., Lees A. J., Wood N. W. (2008) Lancet Neurol. 7, 583–590 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials