Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human - PubMed (original) (raw)
Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human
Krzysztof Kuchta et al. Nucleic Acids Res. 2009 Dec.
Abstract
This article presents a comprehensive review of large and highly diverse superfamily of nucleotidyltransferase fold proteins by providing a global picture about their evolutionary history, sequence-structure diversity and fulfilled functional roles. Using top-of-the-line homology detection method combined with transitive searches and fold recognition, we revised the realm of these superfamily in numerous databases of catalogued protein families and structures, and identified 10 new families of nucleotidyltransferase fold. These families include hundreds of previously uncharacterized and various poorly annotated proteins such as Fukutin/LICD, NFAT, FAM46, Mab-21 and NRAP. Some of these proteins seem to play novel important roles, not observed before for this superfamily, such as regulation of gene expression or choline incorporation into cell membrane. Importantly, within newly detected families we identified 25 novel superfamily members in human genome. Among these newly assigned members are proteins known to be involved in congenital muscular dystrophy, neurological diseases and retinal pigmentosa what sheds some new light on the molecular background of these genetic disorders. Twelve of new human nucleotidyltransferase fold proteins belong to Mab-21 family known to be involved in organogenesis and development. The determination of specific biological functions of these newly detected proteins remains a challenging task.
Figures
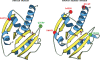
Figure 1.
NTase fold superfamily structures. An example of NTase fold structure is presented together with the observed variation of secondary structure elements and additional domains in representative superfamily proteins of known structure. Positions of conserved active site motifs involved in catalysis ([DE]h[DE]h, h[DE]h) and substrate binding (hG[GS]) are marked above corresponding secondary structure elements. Side-chains of critical motifs residues (aspartates and serine) are shown as balls and sticks. Suffixes ‘_CT’ and ‘_NC’ refer to catalytic and non-catalytic NTase domains.
Figure 2.
Multiple sequence alignment for NTase fold superfamily. Only conserved regions of the fold core are shown for representative proteins of known structure and single representative sequences for groups lacking experimentally solved structure. Sequences are labeled according to PDB code or NCBI gene identification (gi) number and the number of group they belong to (XVII–XXVI, newly detected NTase fold proteins). Multiple NTase fold domains occurring in some proteins are denoted by consecutive roman numbers. The numbers of excluded residues are specified in square brackets. Conservation of residues is denoted with the following scheme: uncharged, highlighted in yellow; small, letters in red; critical active site aspartates/glutamates, highlighted in black. Locations of secondary structure elements (E, β-strand; H, α-helix) are marked above sequences.
Figure 3.
Connectivity network for NTase fold superfamily. Groups of closely related proteins that can be linked by both PSI-BLAST and RPS-BLAST are surrounded by black (known members of NTase fold superfamily) and red (newly detected) circles. Detected Meta-BASIC connections between PFAM, KOG, COG families, PDB structures and human proteins are shown as lines colored according to the Meta-BASIC score: pink, confident scores above 40; grey, below threshold scores between 28 and 40. For each group short description and taxonomy distribution is provided.
Figure 4.
NTase fold proteins in human. Locations of genes encoding NTase fold proteins in human genome are marked with triangles, while rectangles correspond to known genetic diseases associated with mutations found in some of these genes. ENSEMBL identifiers are provided for each protein in blue and red boxes that correspond to known and newly identified groups in our classification, respectively.
Figure 5.
Genetic mutations in Fukutin and Fukutin related protein connected with congenital muscular dystrophy. Positions of single point mutations found in genes encoding Fukutin (in Fukuyama-type congenital muscular dystrophy, FCMD) and Fukutin related protein (in congenital muscular dystrophy type 1C, MDC1C) are shown as spheres on the respective 3D models of their structure. Mutations denoted with green spheres are clustered in the area of the active site and probably impede catalysis or substrate binding. Red spheres represent mutations grouped on the opposite side of the protein surface where they may impair protein–protein or protein–membrane interactions.
Similar articles
- Realm of PD-(D/E)XK nuclease superfamily revisited: detection of novel families with modified transitive meta profile searches.
Knizewski L, Kinch LN, Grishin NV, Rychlewski L, Ginalski K. Knizewski L, et al. BMC Struct Biol. 2007 Jun 20;7:40. doi: 10.1186/1472-6807-7-40. BMC Struct Biol. 2007. PMID: 17584917 Free PMC article. - DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily.
Holm L, Sander C. Holm L, et al. Trends Biochem Sci. 1995 Sep;20(9):345-7. doi: 10.1016/s0968-0004(00)89071-4. Trends Biochem Sci. 1995. PMID: 7482698 No abstract available. - DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history.
Aravind L, Koonin EV. Aravind L, et al. Nucleic Acids Res. 1999 Apr 1;27(7):1609-18. doi: 10.1093/nar/27.7.1609. Nucleic Acids Res. 1999. PMID: 10075991 Free PMC article. - [Fukutin, a novel protein product responsible for Fukuyama-type congenital muscular dystrophy].
Toda T. Toda T. Seikagaku. 1999 Jan;71(1):55-61. Seikagaku. 1999. PMID: 10067123 Review. Japanese. No abstract available. - Unification of protein families.
Holm L. Holm L. Curr Opin Struct Biol. 1998 Jun;8(3):372-9. doi: 10.1016/s0959-440x(98)80072-9. Curr Opin Struct Biol. 1998. PMID: 9666334 Review.
Cited by
- TENT5A mediates the cancer-inhibiting effects of EGR1 by suppressing the protein stability of RPL35 in hepatocellular carcinoma.
Min X, Lin F, Zhao X, Yu J, Ge C, Zhang S, Li X, Zhao F, Chen T, Tian H, Yan M, Li J, Li H. Min X, et al. Cell Oncol (Dordr). 2024 Nov 21. doi: 10.1007/s13402-024-01014-9. Online ahead of print. Cell Oncol (Dordr). 2024. PMID: 39570560 - Inducible auto-phosphorylation regulates a widespread family of nucleotidyltransferase toxins.
Arrowsmith TJ, Xu X, Xu S, Usher B, Stokes P, Guest M, Bronowska AK, Genevaux P, Blower TR. Arrowsmith TJ, et al. Nat Commun. 2024 Sep 4;15(1):7719. doi: 10.1038/s41467-024-51934-1. Nat Commun. 2024. PMID: 39231966 Free PMC article. - In Vitro Cleavage Assay to Characterize DENV NS2B3 Antagonism of cGAS.
Bhattacharya M, Bhowmik D, Yin Q. Bhattacharya M, et al. Methods Mol Biol. 2025;2854:153-170. doi: 10.1007/978-1-0716-4108-8_16. Methods Mol Biol. 2025. PMID: 39192127 - Transition transferases prime bacterial capsule polymerization.
Litschko C, Di Domenico V, Schulze J, Li S, Ovchinnikova OG, Voskuilen T, Bethe A, Cifuente JO, Marina A, Budde I, Mast TA, Sulewska M, Berger M, Buettner FFR, Lowary TL, Whitfield C, Codée JDC, Schubert M, Guerin ME, Fiebig T. Litschko C, et al. Nat Chem Biol. 2025 Jan;21(1):120-130. doi: 10.1038/s41589-024-01664-8. Epub 2024 Jul 1. Nat Chem Biol. 2025. PMID: 38951648 Free PMC article. - Conservation and similarity of bacterial and eukaryotic innate immunity.
Ledvina HE, Whiteley AT. Ledvina HE, et al. Nat Rev Microbiol. 2024 Jul;22(7):420-434. doi: 10.1038/s41579-024-01017-1. Epub 2024 Feb 28. Nat Rev Microbiol. 2024. PMID: 38418927 Free PMC article. Review.
References
- Rogozin IB, Aravind L, Koonin EV. Differential action of natural selection on the N and C-terminal domains of 2′-5′ oligoadenylate synthetases and the potential nuclease function of the C-terminal domain. J. Mol. Biol. 2003;326:1449–1461. - PubMed
- Tomita K, Fukai S, Ishitani R, Ueda T, Takeuchi N, Vassylyev DG, Nureki O. Structural basis for template-independent RNA polymerization. Nature. 2004;430:700–704. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases