Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain - PubMed (original) (raw)
Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain
Junji Otani et al. EMBO Rep. 2009 Nov.
Abstract
DNMT3 proteins are de novo DNA methyltransferases that are responsible for the establishment of DNA methylation patterns in mammalian genomes. Here, we have determined the crystal structures of the ATRX-DNMT3-DNMT3L (ADD) domain of DNMT3A in an unliganded form and in a complex with the amino-terminal tail of histone H3. Combined with the results of biochemical analysis, the complex structure indicates that DNMT3A recognizes the unmethylated state of lysine 4 in histone H3. This finding indicates that the recruitment of DNMT3A onto chromatin, and thereby de novo DNA methylation, is mediated by recognition of the histone modification state by its ADD domain. Furthermore, our biochemical and nuclear magnetic resonance data show mutually exclusive binding of the ADD domain of DNMT3A and the chromodomain of heterochromatin protein 1alpha to the H3 tail. These results indicate that de novo DNA methylation by DNMT3A requires the alteration of chromatin structure.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
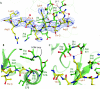
Overall structures of ADD3A in the unliganded and H3-bound forms. (A) Ribbon representation of the ligand-free ADD3A. Three zinc ions and cysteine residues forming CxxC motifs are shown as magenta spheres and multicoloured stick models, respectively. GATA1-like and PHD fingers are shown in orange and green, respectively. The green dotted line represents the disordered H3N loop. (B) Crystal structure of ADD3A (green) bound to the H3 tail (yellow). H3 peptide (residues 1–9) is shown as a ball-and-stick model. The disordered linker peptide that connects the carboxy-terminus of the peptide and the amino-terminus of ADD3A is represented by a yellow dotted line. ADD3A, ADD domain of DNMT3A; GATA1, GATA binding protein 1; PHD, plant homeodomain.
Figure 2
Calorimetric study of the binding of ADD3A to H31–19 peptides containing non-methylated (A), dimethylated (B) or trimethylated (C) H3K4 analogues. Δ_S_, Δ_H_ and _K_D values are the means of two experiments that used different peptide and protein concentrations. H31–19, amino-terminal 19 amino acids of histone H3.
Figure 3
Recognition of non-methylated H3K4 by ADD3A. Red dotted lines indicate hydrogen bonds (<3.3 Å) between H3 and ADD3A residues. ADD3A and H3 residues are represented as ball-and-stick models in green and yellow, respectively. (A) H3 peptide is shown with a _F_o−_F_c omit map contoured at 2.0 σ. (B) A close-up view of the recognition of the amino-terminus of the H3 tail by ADD3A. (C) A close-up view of the Lys 4 binding pocket of ADD3A. ADD3A, ADD domain of DNMT3A.
Figure 4
Competitive binding of ADD3A and CDHP1α to the H3 tail. (A) GST pulldown assay analysed by SDS–PAGE and CBB staining. The GST–CDHP1α bound to H3 K9me2 peptide in the absence of ADD3A (lane 5) and was competed out by adding ADD3A (lanes 6–9). (B) Reciprocal experiment of (A) using GST–ADD3A, the H3 K9me3 peptide and CDHP1α. (C) 1H–15N HSQC spectra of the 15N-labelled CDHP1α in the ligand-free (green) and H3 K9me3-bound (red) forms are superimposed (left panel). The CDHP1α–H3 K9me3 complex was titrated with unlabelled ADD3A (right panel). The cross-peak positions of 15N-labelled CDHP1α shifted from those of the H3-bound form to those of the free form during the titration. Selected spectral regions (i)–(iv) are magnified. (D,E) Reciprocally, 15N-labelled ADD3A in complex with H3K9me2 (D) or H3K9me3 peptide (E) was titrated by unlabelled CDHP1α. ADD, ATRX–DNMT3–DNMT3L; ADD3A, ADD domain of DNMT3A; CBB, coomassie brilliant blue; CDHP1α, chromodomain of HP1α; GST, glutathione-_S-_transferase; HP1α, heterochromatin protein 1α; SDS–PAGE, sodium dodecyl sulphate–polyacrylamide gel electrophoresis.
Similar articles
- Structural insight into autoinhibition and histone H3-induced activation of DNMT3A.
Guo X, Wang L, Li J, Ding Z, Xiao J, Yin X, He S, Shi P, Dong L, Li G, Tian C, Wang J, Cong Y, Xu Y. Guo X, et al. Nature. 2015 Jan 29;517(7536):640-4. doi: 10.1038/nature13899. Epub 2014 Nov 10. Nature. 2015. PMID: 25383530 - DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA.
Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. Ooi SK, et al. Nature. 2007 Aug 9;448(7154):714-7. doi: 10.1038/nature05987. Nature. 2007. PMID: 17687327 Free PMC article. - Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation.
Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Jia D, et al. Nature. 2007 Sep 13;449(7159):248-51. doi: 10.1038/nature06146. Epub 2007 Aug 22. Nature. 2007. PMID: 17713477 Free PMC article. - Molecular coupling of DNA methylation and histone methylation.
Hashimoto H, Vertino PM, Cheng X. Hashimoto H, et al. Epigenomics. 2010 Oct;2(5):657-69. doi: 10.2217/epi.10.44. Epigenomics. 2010. PMID: 21339843 Free PMC article. Review. - Molecular and enzymatic profiles of mammalian DNA methyltransferases: structures and targets for drugs.
Xu F, Mao C, Ding Y, Rui C, Wu L, Shi A, Zhang H, Zhang L, Xu Z. Xu F, et al. Curr Med Chem. 2010;17(33):4052-71. doi: 10.2174/092986710793205372. Curr Med Chem. 2010. PMID: 20939822 Free PMC article. Review.
Cited by
- The interplay between DNA and histone methylation: molecular mechanisms and disease implications.
Li Y, Chen X, Lu C. Li Y, et al. EMBO Rep. 2021 May 5;22(5):e51803. doi: 10.15252/embr.202051803. Epub 2021 Apr 12. EMBO Rep. 2021. PMID: 33844406 Free PMC article. Review. - Distinctive aspects of the placental epigenome and theories as to how they arise.
Pastor WA, Kwon SY. Pastor WA, et al. Cell Mol Life Sci. 2022 Oct 26;79(11):569. doi: 10.1007/s00018-022-04568-9. Cell Mol Life Sci. 2022. PMID: 36287261 Free PMC article. Review. - Reversible Regulation of Promoter and Enhancer Histone Landscape by DNA Methylation in Mouse Embryonic Stem Cells.
King AD, Huang K, Rubbi L, Liu S, Wang CY, Wang Y, Pellegrini M, Fan G. King AD, et al. Cell Rep. 2016 Sep 27;17(1):289-302. doi: 10.1016/j.celrep.2016.08.083. Cell Rep. 2016. PMID: 27681438 Free PMC article. - Expression level and immunolocalization of de novo methyltransferase 3 protein (TuDNMT3) in adult females and males of the two-spotted spider mite, Tetranychus urticae.
Yang SX, Guo C, Zhang YK, Sun JT, Hong XY. Yang SX, et al. Exp Appl Acarol. 2015 Nov;67(3):381-92. doi: 10.1007/s10493-015-9957-5. Epub 2015 Aug 6. Exp Appl Acarol. 2015. PMID: 26246190 - Chromatin-dependent allosteric regulation of DNMT3A activity by MeCP2.
Rajavelu A, Lungu C, Emperle M, Dukatz M, Bröhm A, Broche J, Hanelt I, Parsa E, Schiffers S, Karnik R, Meissner A, Carell T, Rathert P, Jurkowska RZ, Jeltsch A. Rajavelu A, et al. Nucleic Acids Res. 2018 Sep 28;46(17):9044-9056. doi: 10.1093/nar/gky715. Nucleic Acids Res. 2018. PMID: 30102379 Free PMC article.
References
- Ayoub N, Jeyasekharan A, Bernal J, Venkitaraman A (2008) HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature 453: 682–686 - PubMed
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 - PubMed
- Cryderman D, Cuaycong M, Elgin S, Wallrath L (1998) Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma 107: 277–285 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases