Improved reproducibility of reverse-phase protein microarrays using array microenvironment normalization - PubMed (original) (raw)
Improved reproducibility of reverse-phase protein microarrays using array microenvironment normalization
Troy Anderson et al. Proteomics. 2009 Dec.
Abstract
We introduce a novel experimental methodology for the reverse-phase protein microarray platform which reduces the typical measurement CV as much as 70%. The methodology, referred to as array microenvironment normalization, increases the statistical power of the platform. In the experiment, it enabled the detection of a 1.1-fold shift in prostate specific antigen concentration using approximately six technical replicates rather than the 37 replicates previously required. The improved reproducibility and statistical power should facilitate clinical implementation of the platform.
Conflict of interest statement
Conflict of interest: Authors (excluding Dr. Raimond Winslow) have affiliation with Theranostic Health Inc. which is commercializing the RPMA technology.
Figures
Figure 1
RPMA with AMN experimental design. Top is the array 1 total protein stain. Bottom is a rendering of the “checkerboard” control layout used to carry out AMN for each sample spot.
Figure 2
Linearity and reproducibility improvement with AMN demonstrated for array 1. (A) Scatter plot of TPN relative intensity (no AMN) for each sample of array 1 versus the known PSA concentration. (B) Scatter plot of AMN relative intensity for each sample of array 1 versus the known PSA concentration. (C) Scatter plot of the AMN relative intensity versus TPN relative intensity for samples of array 1 which differed in PSA concentration by 1.4-fold. (D) Same as (C) except displays samples differing in PSA concentration by 1.1-fold for array 1. The spread in the data is always much greater before AMN is implemented.
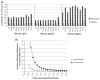
Figure 3
AMN improves the average coefficient of variance by 72% which results in an increased power to detect shifts in expression with fewer replicates. (A) Bar plots of measured coefficient of variance for the 32 replicates of each sample containing the specified amount of PSA for arrays 1, 2 and 3 both with and without AMN. (B) Power curve displaying the minimum number of technical replicates needed to detect the fold change specified in the _x_-axis with p<0.05 and power >0.80 using a _t_-test when the CV=5 or 15% (representative of the CV with and without AMN).
Similar articles
- Reverse-phase versus sandwich antibody microarray, technical comparison from a clinical perspective.
Järås K, Ressine A, Nilsson E, Malm J, Marko-Varga G, Lilja H, Laurell T. Järås K, et al. Anal Chem. 2007 Aug 1;79(15):5817-25. doi: 10.1021/ac0709955. Epub 2007 Jul 3. Anal Chem. 2007. PMID: 17605470 - Robust-linear-model normalization to reduce technical variability in functional protein microarrays.
Sboner A, Karpikov A, Chen G, Smith M, Mattoon D, Freeman-Cook L, Schweitzer B, Gerstein MB. Sboner A, et al. J Proteome Res. 2009 Dec;8(12):5451-64. doi: 10.1021/pr900412k. J Proteome Res. 2009. PMID: 19817483 - PMA: Protein Microarray Analyser, a user-friendly tool for data processing and normalization.
Da Gama Duarte J, Goosen RW, Lawry PJ, Blackburn JM. Da Gama Duarte J, et al. BMC Res Notes. 2018 Feb 27;11(1):156. doi: 10.1186/s13104-018-3266-0. BMC Res Notes. 2018. PMID: 29482592 Free PMC article. - Why 3-D? Gel-based microarrays in proteomics.
Rubina AY, Kolchinsky A, Makarov AA, Zasedatelev AS. Rubina AY, et al. Proteomics. 2008 Feb;8(4):817-31. doi: 10.1002/pmic.200700629. Proteomics. 2008. PMID: 18214844 Review. - Extrapolating traditional DNA microarray statistics to tiling and protein microarray technologies.
Royce TE, Rozowsky JS, Luscombe NM, Emanuelsson O, Yu H, Zhu X, Snyder M, Gerstein MB. Royce TE, et al. Methods Enzymol. 2006;411:282-311. doi: 10.1016/S0076-6879(06)11015-0. Methods Enzymol. 2006. PMID: 16939796 Review.
Cited by
- Integrated imaging instrument for self-calibrated fluorescence protein microarrays.
Reddington AP, Monroe MR, Ünlü MS. Reddington AP, et al. Rev Sci Instrum. 2013 Oct;84(10):103702. doi: 10.1063/1.4823790. Rev Sci Instrum. 2013. PMID: 24182114 Free PMC article. - NormaCurve: a SuperCurve-based method that simultaneously quantifies and normalizes reverse phase protein array data.
Troncale S, Barbet A, Coulibaly L, Henry E, He B, Barillot E, Dubois T, Hupé P, de Koning L. Troncale S, et al. PLoS One. 2012;7(6):e38686. doi: 10.1371/journal.pone.0038686. Epub 2012 Jun 28. PLoS One. 2012. PMID: 22761696 Free PMC article. - Reverse phase protein microarrays advance to use in clinical trials.
Mueller C, Liotta LA, Espina V. Mueller C, et al. Mol Oncol. 2010 Dec;4(6):461-81. doi: 10.1016/j.molonc.2010.09.003. Epub 2010 Oct 16. Mol Oncol. 2010. PMID: 20974554 Free PMC article. Review. - Analysis of Reverse Phase Protein Array Data: From Experimental Design towards Targeted Biomarker Discovery.
Wachter A, Bernhardt S, Beissbarth T, Korf U. Wachter A, et al. Microarrays (Basel). 2015 Nov 3;4(4):520-39. doi: 10.3390/microarrays4040520. Microarrays (Basel). 2015. PMID: 27600238 Free PMC article. Review. - Probe-target hybridization depends on spatial uniformity of initial concentration condition across large-format chips.
Geldert A, Huang H, Herr AE. Geldert A, et al. Sci Rep. 2020 May 29;10(1):8768. doi: 10.1038/s41598-020-65563-3. Sci Rep. 2020. PMID: 32472029 Free PMC article.
References
- Kricka LJ, Master SR. Validation and quality control of protein microarray-based analytical methods. Mol Biotechnol. 2008;38:19–31. - PubMed
- Spurrier B, Ramalingam S, Nishizuka S. Reverse-phase protein lysate microarrays for cell signaling analysis. Nat Protoc. 2008;3:1796–1808. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical