Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes - PubMed (original) (raw)
. 2009 Oct 22;6(4):367-80.
doi: 10.1016/j.chom.2009.09.005.
Dorothee Dormann, Randy Albrecht, Jörn Dengjel, Tania Torossi, Patrick C Rämer, Monica Lee, Till Strowig, Frida Arrey, Gina Conenello, Marc Pypaert, Jens Andersen, Adolfo García-Sastre, Christian Münz
Affiliations
- PMID: 19837376
- PMCID: PMC2774833
- DOI: 10.1016/j.chom.2009.09.005
Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes
Monique Gannagé et al. Cell Host Microbe. 2009.
Abstract
Influenza A virus is an important human pathogen causing significant morbidity and mortality every year and threatening the human population with epidemics and pandemics. Therefore, it is important to understand the biology of this virus to develop strategies to control its pathogenicity. Here, we demonstrate that influenza A virus inhibits macroautophagy, a cellular process known to be manipulated by diverse pathogens. Influenza A virus infection causes accumulation of autophagosomes by blocking their fusion with lysosomes, and one viral protein, matrix protein 2, is necessary and sufficient for this inhibition of autophagosome degradation. Macroautophagy inhibition by matrix protein 2 compromises survival of influenza virus-infected cells but does not influence viral replication. We propose that influenza A virus, which also encodes proapoptotic proteins, is able to determine the death of its host cell by inducing apoptosis and also by blocking macroautophagy.
Figures
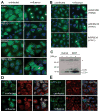
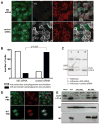
Figure 1. Classical autophagosomes accumulation after influenza A virus infection
(A) Stably GFP-Atg8/LC3 transfected epithelial cells lines A549, MLE-12, MDAMC and HaCat were infected with influenza A/Aichi/68 virus and were analyzed for GFP-Atg8/LC3 positive autophagosome accumulation 24h later by fluorescence microscopy. DAPI was used to stain nuclear DNA. Scale bar: 30 μm. (B) GFP-Atg8/LC3 transfected A549 human lung epithelial cells were infected with different influenza A virus strains (A/Aichi/68, A/WSN/33 and A/PR8/34) and analyzed after 24h. One of three experiments is shown. Scale bar: 30 μm. (C) The human epithelial cell lines HaCat and 293T were infected with influenza A/Aichi/68 virus at a MOI of 0.1. 24h post-infection, lysates of uninfected and infected cells were analyzed by Western blot analysis for Atg8/LC3. Unconjugated (LC3-I) and lipidated Atg8/LC3 (LC3-II) can be distinguished by there apparent molecular weights of 18kD and 16kD, respectively. The asterisk (*) marks an unspecific band, detected by our Atg8/LC3 antiserum. One of three experiments is shown. 24h after influenza A virus infection co-localization of GFP-ATG8/LC3 and the classical macroautophagy substrates poly-ubiquitinated proteins (D) and p62/sequestosome 1 (E) was analyzed by immune fluorescence microscopy. Nuclear DNA was stained with DAPI. Scale bar: 20 μm. One of three experiments is shown.
Figure 2. Autophagosomes do not fuse with acidified proteolytic lysosomes in influenza infected cells
(A) Live cell imaging was performed to analyze co-localization of lysotracker stained acidified vesicles and GFP-Atg8/LC3 positive autophagosomes in uninfected and influenza A virus infected cells. Scale bar: 15 μm. Representative still images of two independent experiments are shown. (B) Furthermore, fusion of autophagosomes with lysosomes was analyzed as co-localization of the autophagosomes marker GFP-Atg8/LC3 with the lysosome marker LAMP1. Nuclear DNA was stained with DAPI and infected cells with matrix protein 1 (M1) specific antibodies. Scale bar: 30 μm. One of three experiments is shown. (C) GFP-Atg8/LC3 transfected A549 human lung epithelial and MDAMC human breast carcinoma cells were infected with influenza A virus at a MOI of 0.4 for 24h. Then the indicated samples were treated for 6h with the lysosomal inhibitor chloroquine (CQ), and lipidated endogenous Atg8/LC3-II and lipidated transfected GFP-Atg8/LC3-II content was analyzed by Atg8/LC3 specific Western blot analysis. One of three experiments is shown. (D) The tandem reporter construct mRFP-GFP-Atg8/LC3 was transiently transfected into uninfected or influenza A virus infected A549 human lung epithelial cells. GFP (sensitive to acidification and lysosomal degradation) and RFP fluorescence (insensitive to acidification and lysosomal degradation) of the reporter construct were analyzed by fluorescence microscopy. DAPI was used to stain nuclear DNA. Scale bar: 20 μm. One of three experiments is shown.
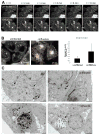
Figure 3. Mobility and ultrastructure of autophagosomes in influenza A virus infected cells
(A) Live cell imaging of GFP-Atg8/LC3 transfected A549 human lung epithelial cells was performed to analyze autophagosome mobility. Still images of uninfected and influenza A virus infected cells taken at the indicated timepoints of live cell imaging document the immobility of the accumulated large perinuclear autophagosomes. (B) Autphagosome tracking in uninfected and influenza A infected cells. Left: Vesicle tracks are displayed as color coded tracks, indicating the distance traveled by one vesicle during the indicated observation period (t=0 in blue, t=endpoint in white). Right: Average speed of autophagosomes was determined in three independent fields and summarized. One of three experiments is shown. Error bars indicate standard deviation. (C) Electron micrographs of uninfected and influenza A virus infected A549 human lung epithelial cells. Small autophagic structures (black arrows, upper row) were detected in uninfected cells, whereas large autophagic structures (black arrows, lower row) were visible in infected cells. Scale bar: 2 μm. One of three experiments is shown.
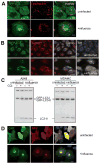
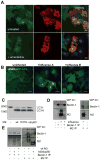
Figure 4. Influenza A virus matrix protein 2 (M2) blocks autophagosome fusion with lysosomes
(A) GFP-Atg8/LC3 transfected A549 human lung epithelial cells were transfected with expression plasmids encoding either FLAG tagged M2 or NP of influenza A/WSN/33 virus. 24 hours post transfection GFP-ATG8/LC3 positive autophagosome accumulation was analyzed by fluorescence microscopy. Influenza M2 specific antibodies were used for M2 detection, and an anti-Flag antibody for NP detection. Scale bar: 80 μm. One of three experiments is shown. (B) Left: Western Blot analysis of lipidated endogenous Atg8/LC3-II accumulation in A549 cells transfected with M2 or NP. Right: Western blot analysis of M2 and NP expression in transfected cells, using anti-FLAG tag staining. One of three experiments is shown. (C) Colocalization analysis of M2 (red fluorescence) and GFP-LC3 (green fluorescence) in infected A549 cells: Serial optical sections, (0.2 um; 40 sections) were acquired and images were then deconvoluted using Huygens software. Pearson coefficients were calculated using the Imaris 6.3.0 software. White fluorescence represents colocalization. Scale bar: 10μm. One representative of 36 analyzed cells is shown. (D). Mass spectrometric analysis of density gradient fractions (1–6) of influenza A virus infected A549 cells content upon influenza infection in A549 cells. Cell organelle marker protein distribution in these fractions was determined by mass spectrometric sequencing of protein fragments. Influenza A virus M2 distribution follows the distribution of autophagosome marker protein (right panel), but not the distribution pattern of mitochondria and endoplasmic reticulum (ER) proteins. One of four experiments is shown.
Figure 5. Loss of influenza A virus matrix protein 2 (M2) expression during infection prevents autophagosome accumulation
(A) GFP-Atg8/LC3 transfected A549 cells were transfected with M2 specific siRNA or a control siRNA, and analyzed 24 hours after infection by fluorescence microscopy. In order to monitor influenza A virus infection and M2 expression, co-staining for NP and M2 was performed. Scale bar: 30 μm. One of three experiments is shown. (B) Summary of perinuclear autophagosomes accumulation in influenza A virus infected cells with and without M2 specific RNA silencing of three independent experiments. In each experiment at least 150 cells were analyzed per condition. Statistical analysis was performed by applying Pearson’s chi-squared test. (C) Western blot analysis of lipidated Atg8/LC3-II accumulation in influenza infected A549 cells transfected with M2 siRNA or control siRNA. One of three experiments is shown. (D) GFP-Atg8/LC3 transfected A549 cells at 24 hours post infection with M2 deficient M2- PR8) or wild-type influenza A virus (WT PR8). Infection was visualized by staining for NP. Scale bar: 100 μm. (E) Western blot analysis of Atg8/LC3-II, M2 and NP expression in influenza A virus infected A549 cells using the two recombinant influenza A viruses (M2- PR8 and WT PR8) at MOIs of 1 and 10, and influenza A/X:31 virus as a control. One of five experiments is shown.
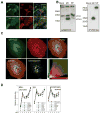
Figure 6. The proton channel function of M2 is not involved in autophagosomes accumulation
(A) GFP-Atg8/LC3 transfected A549 human lung epithelial cells were infected with influenza A virus and incubated one hour later with and without amantadine at a concentration of 50μM. Cells were stained with M2 antibody and analyzed for GFP-Atg8/LC3 positive autophagosome accumulation by fluorescence microscopy. Nuclear DNA was stained with DAPI. Scale bar: 100 μm. Representative images of one out of three experiments are shown. (B) GFP-Atg8/LC3 transfected A549 cells were infected with influenza A or B and analyzed by fluorescence microscopy for autophagosome accumulation. DAPI was used to stain nuclear DNA. Scale bar: 100 μm. One of three experiments is shown. (C) A549 cells were transiently transfected with M2 wild type and M2 mutants (M2 H37G and M2 stop60) encoding plasmids. 24h post transfection Atg8/LC3-II accumulation was analyzed by Western blot analysis. One of three experiments is shown. (D) A549 cells were infected with X:31 virus, 24 hour later cell lysates from infected and non infected cells were immunoprecipitated using Atg6/Beclin-1 specific (left) or M2 specific antibodies (right). Western blotting was performed for Atg6/Beclin-1 and influenza A virus M2 detection. One of 3 experiments is shown. (E) A549 cells were transfected with full-length (wt) or the 60 N-terminal amino acids of M2 (stop60). Transfected or non-transfected cells were immunoprecipitated using Atg6/Beclin-1 or M2 specific antibodies (Atg6/Beclin-1 IP: left panel, M2 IP: right panel). Western blotting was performed for Atg6/Beclin-1 (top) and M2 (bottom) detection. * indicates immunoglobulin heavy chain of the antibodies used for immunoprecipitation. One of three experiments is shown.
Figure 7. Macroautophagy inhibition enhances cell death after influenza A virus infection
(A) Wild-type (Atg5+/+) and macroautophagy deficient (ATG5−/−) MEFs were infected with the indicated doses of influenza A virus (hemagglutinin units [HAU] per 106 cells), and analyzed 24 hours later by flow cytometry. Staining for M2, 7-AAD and annexin V was performed. Cells were gated on M2 positive cells, and apoptosis (annexin V positive) and secondary necrosis (annexin V and 7-AAD positive) cells were quantified. The percentage of double positive cells (annexin V and 7-AAD positive) is significantly higher in ATG5−/−cells (paired t test p=0.005). One of five experiments is shown. (B) Cell death induction was analyzed as in (A), but with one constant influenza A virus infection dose (1200HAU/106 cells) at the indicated time points. One of three experiments is shown. (C). A549 cells were infected with the M2 deficient or the wild-type recombinant virus at an MOI of 1 and cell death was analyzed at 24 hours post infection. Cells were gated on infected cells, (anti-H1N1 FITC) and apoptosis (annexin V positive) and secondary necrosis (annexin V and 7-AAD positive) cells were quantified. (D) Composite data on five independent experiments comparing apoptosis (Annexin V+) and secondary necrosis (7AAD+ Annexin V+) after M2 deficient (M2- PR8) and wild-type (WT PR8) influenza A virus infection of A549 cells.
Comment in
- Autophagy, apoptosis, and the influenza virus M2 protein.
Rossman JS, Lamb RA. Rossman JS, et al. Cell Host Microbe. 2009 Oct 22;6(4):299-300. doi: 10.1016/j.chom.2009.09.009. Cell Host Microbe. 2009. PMID: 19837369 Review.
Similar articles
- Autophagy, apoptosis, and the influenza virus M2 protein.
Rossman JS, Lamb RA. Rossman JS, et al. Cell Host Microbe. 2009 Oct 22;6(4):299-300. doi: 10.1016/j.chom.2009.09.009. Cell Host Microbe. 2009. PMID: 19837369 Review. - Influenza virus M2 targets cystic fibrosis transmembrane conductance regulator for lysosomal degradation during viral infection.
Londino JD, Lazrak A, Noah JW, Aggarwal S, Bali V, Woodworth BA, Bebok Z, Matalon S. Londino JD, et al. FASEB J. 2015 Jul;29(7):2712-25. doi: 10.1096/fj.14-268755. Epub 2015 Mar 20. FASEB J. 2015. PMID: 25795456 Free PMC article. - Proton Channel Activity of Influenza A Virus Matrix Protein 2 Contributes to Autophagy Arrest.
Ren Y, Li C, Feng L, Pan W, Li L, Wang Q, Li J, Li N, Han L, Zheng X, Niu X, Sun C, Chen L. Ren Y, et al. J Virol. 2015 Oct 14;90(1):591-8. doi: 10.1128/JVI.00576-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26468520 Free PMC article. - Phosphoprotein of human parainfluenza virus type 3 blocks autophagosome-lysosome fusion to increase virus production.
Ding B, Zhang G, Yang X, Zhang S, Chen L, Yan Q, Xu M, Banerjee AK, Chen M. Ding B, et al. Cell Host Microbe. 2014 May 14;15(5):564-77. doi: 10.1016/j.chom.2014.04.004. Cell Host Microbe. 2014. PMID: 24832451 - Regulation of Autophagosome-Lysosome Fusion by Human Viral Infections.
Ke PY. Ke PY. Pathogens. 2024 Mar 20;13(3):266. doi: 10.3390/pathogens13030266. Pathogens. 2024. PMID: 38535609 Free PMC article. Review.
Cited by
- Viral Membrane Channels: Role and Function in the Virus Life Cycle.
Sze CW, Tan YJ. Sze CW, et al. Viruses. 2015 Jun 23;7(6):3261-84. doi: 10.3390/v7062771. Viruses. 2015. PMID: 26110585 Free PMC article. Review. - Viruses and the autophagy pathway.
Jackson WT. Jackson WT. Virology. 2015 May;479-480:450-6. doi: 10.1016/j.virol.2015.03.042. Epub 2015 Apr 6. Virology. 2015. PMID: 25858140 Free PMC article. Review. - The Emerging Roles of Viroporins in ER Stress Response and Autophagy Induction during Virus Infection.
Fung TS, Torres J, Liu DX. Fung TS, et al. Viruses. 2015 Jun 4;7(6):2834-57. doi: 10.3390/v7062749. Viruses. 2015. PMID: 26053926 Free PMC article. Review. - Structure of the extracellular domain of matrix protein 2 of influenza A virus in complex with a protective monoclonal antibody.
Cho KJ, Schepens B, Seok JH, Kim S, Roose K, Lee JH, Gallardo R, Van Hamme E, Schymkowitz J, Rousseau F, Fiers W, Saelens X, Kim KH. Cho KJ, et al. J Virol. 2015 Apr;89(7):3700-11. doi: 10.1128/JVI.02576-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609808 Free PMC article. - Autophagy and viruses: adversaries or allies?
Dong X, Levine B. Dong X, et al. J Innate Immun. 2013;5(5):480-93. doi: 10.1159/000346388. Epub 2013 Jan 31. J Innate Immun. 2013. PMID: 23391695 Free PMC article. Review.
References
- Baskin CR, Bielefeldt-Ohmann H, Tumpey TM, Sabourin PJ, Long JP, Garcia-Sastre A, Tolnay AE, Albrecht R, Pyles JA, Olson PH, et al. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc Natl Acad Sci U S A. 2009;106:3455–3460. - PMC - PubMed
- Bender A, Albert M, Reddy A, Feldman M, Sauter B, Kaplan G, Hellman W, Bhardwaj N. The distinctive features of influenza virus infection of dendritic cells. Immunobiology. 1998;198:552–567. - PubMed
- Brydon EW, Smith H, Sweet C. Influenza A virus-induced apoptosis in bronchiolar epithelial (NCI-H292) cells limits pro-inflammatory cytokine release. J Gen Virol. 2003;84:2389–2400. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U19AI62623/AI/NIAID NIH HHS/United States
- R01 CA108609/CA/NCI NIH HHS/United States
- U54AI57158/AI/NIAID NIH HHS/United States
- R01 CA101741/CA/NCI NIH HHS/United States
- U54 AI057158/AI/NIAID NIH HHS/United States
- R01CA101741/CA/NCI NIH HHS/United States
- U19 AI062623/AI/NIAID NIH HHS/United States
- R01CA108609/CA/NCI NIH HHS/United States
- HHSN266200700010C/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous