DNA damage signalling prevents deleterious telomere addition at DNA breaks - PubMed (original) (raw)
. 2009 Nov;11(11):1383-6.
doi: 10.1038/ncb1985. Epub 2009 Oct 18.
Affiliations
- PMID: 19838171
- PMCID: PMC2806817
- DOI: 10.1038/ncb1985
DNA damage signalling prevents deleterious telomere addition at DNA breaks
Svetlana Makovets et al. Nat Cell Biol. 2009 Nov.
Abstract
The response to DNA damage involves regulation of several essential processes to maximize the accuracy of DNA damage repair and cell survival. Telomerase has the potential to interfere with repair by inappropriately adding telomeres to DNA breaks. It was unknown whether cells modulate telomerase in response to DNA damage to increase the accuracy of repair. Here, we report that telomerase action is regulated as a part of the cellular response to DNA double-strand breaks (DSBs). Using yeast, we show that the main ATR/Mec1 DNA damage signalling pathway regulates telomerase action at DSBs. After DNA damage, MEC1-RAD53-DUN1-dependent phosphorylation of the telomerase inhibitor Pif1 occurs. Using a separation of function PIF1 mutation, we show that this phosphorylation is specifically required for the Pif1-mediated telomerase inhibition that takes place at DNA breaks, but not for that at telomeres. Hence DNA damage signalling down-modulates telomerase action at DNA breaks through Pif1 phosphorylation, thus preventing aberrant healing of broken DNA ends by telomerase. These findings uncover a new regulatory mechanism that coordinates competing DNA end-processing activities and thereby promotes DNA repair accuracy and genome integrity.
Figures
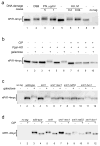
Figure 1
nPif1 is phosphorylated in response to DNA damage in a MEC1-RAD53 dependent manner. In all the panels (a–d), gel mobility of nPif1-4myc either from total protein extracts (panels a and c) or after immunoprecipitation and CIP phosphatase treatments (panels b and d) is analysed by western blotting using an α-myc antibody. a, Analysis of nPif1 gel mobility in response to different DNA damage stimuli. Cells were grown to mid-log phase in rich medium with raffinose. DSBs were induced by addition of galactose (see Methods Summary). Alternatively, either phleomycin (Phl) or hydroxyurea (HU) were added to the concentrations indicated. After 2 h cells were harvested for protein analysis. b, nPif1 is phosphorylated in response to a single DSB induced by the expression of the HO endonuclease from a galactose-inducible promoter. c, nPif1 phosphorylation in response to a single DSB requires MEC1 and RAD53. d, Basal phosphorylation of nPif1 is independent of the DNA damage signalling kinases.
Figure 2
pif1-4A is a separation of function phospho-site mutant, defective in telomerase inhibition specifically at DSBs and not telomeres. a, Mutagenesis based scanning of nPif1 for potential phosphorylation loci involved in its function during DNA damage response. Schematic of the multiple S/T→A substitutions constructed in different nPif1 regions (See Methods for details) and summary of mutant phenotypes (on the right, data presented in Fig. S2). b, Telomere length analysis in PIF1, pif1-m2, pif1-4D, and pif1-4A cells. c, Frequency of 5-FOAR colony formation in PIF1, pif1-m2, pif1-4A, and pif1-4D upon DSB induction. Error bars represent average ± s.d. from four independent measurements for each strain, except pif1-4D (three measurements). Note that the majority of 5-FOAR colonies from PIF1 cells do not represent de novo telomere addition events whereas the majority of the same class of clones from pif1-m2 and pif1-4A do (see the numbers under the graph and Fig. S3). d, Analysis of nPif1-4myc and nPif1-4A-4myc localization to a galactose inducible DSB by chromatin immunoprecipitation. Error bars show average ± s.d. from four independent experiments. e, TLSSAES is phosphorylated in response to a single DSB. Note that the anti-P-Pif1 antibody has weak cross-reactivity with another DNA damage induced phosphorylation site on Pif1 as seen in lane 6 (pif1-4A in galactose). However, this does not affect the conclusiveness of the data as there is a significant signal difference between PIF1 and pif1-4A (compare lanes 4 and 6). The same applies to panels f and g. f, TLSSAES phosphorylation in response to a single DSB requires MEC1, RAD53, and DUN1. g, TLSSAES is phosphorylated in response to DSBs but not in response to nocodazole-induced G2 arrest. h, TLSSAES is phosphorylated in response to DSBs (DSB and Phleomycin, Phl) but not stalled replication forks (hydroxyurea, HU). DNA damage was induced as in Fig. 1. In the panels e–h, samples of immunoprecipitated nPif1-4myc were analyzed by western blotting using an affinity purified rabbit polyclonal antibody raised against VIDFYL(pT)LS(pS)AE (anti-P-Pif1, upper blot on each panel) and then re-probed with anti-myc antibody (lower blot).
Similar articles
- Break-induced replication requires DNA damage-induced phosphorylation of Pif1 and leads to telomere lengthening.
Vasianovich Y, Harrington LA, Makovets S. Vasianovich Y, et al. PLoS Genet. 2014 Oct 16;10(10):e1004679. doi: 10.1371/journal.pgen.1004679. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25329304 Free PMC article. - A sharp Pif1-dependent threshold separates DNA double-strand breaks from critically short telomeres.
Strecker J, Stinus S, Caballero MP, Szilard RK, Chang M, Durocher D. Strecker J, et al. Elife. 2017 Aug 3;6:e23783. doi: 10.7554/eLife.23783. Elife. 2017. PMID: 28826474 Free PMC article. - The regulation of the DNA damage response at telomeres: focus on kinases.
Galli M, Frigerio C, Longhese MP, Clerici M. Galli M, et al. Biochem Soc Trans. 2021 Apr 30;49(2):933-943. doi: 10.1042/BST20200856. Biochem Soc Trans. 2021. PMID: 33769480 Review. - Telomerase is essential to alleviate pif1-induced replication stress at telomeres.
Chang M, Luke B, Kraft C, Li Z, Peter M, Lingner J, Rothstein R. Chang M, et al. Genetics. 2009 Nov;183(3):779-91. doi: 10.1534/genetics.109.107631. Epub 2009 Aug 24. Genetics. 2009. PMID: 19704012 Free PMC article. - Telomeres and DNA damage checkpoints.
Viscardi V, Clerici M, Cartagena-Lirola H, Longhese MP. Viscardi V, et al. Biochimie. 2005 Jul;87(7):613-24. doi: 10.1016/j.biochi.2004.10.022. Epub 2004 Dec 9. Biochimie. 2005. PMID: 15989978 Review.
Cited by
- The telomere syndromes.
Armanios M, Blackburn EH. Armanios M, et al. Nat Rev Genet. 2012 Oct;13(10):693-704. doi: 10.1038/nrg3246. Epub 2012 Sep 11. Nat Rev Genet. 2012. PMID: 22965356 Free PMC article. Review. - Telomeres: structures in need of unwinding.
Paeschke K, McDonald KR, Zakian VA. Paeschke K, et al. FEBS Lett. 2010 Sep 10;584(17):3760-72. doi: 10.1016/j.febslet.2010.07.007. Epub 2010 Jul 14. FEBS Lett. 2010. PMID: 20637196 Free PMC article. Review. - DNA-end capping by the budding yeast transcription factor and subtelomeric binding protein Tbf1.
Ribaud V, Ribeyre C, Damay P, Shore D. Ribaud V, et al. EMBO J. 2012 Jan 4;31(1):138-49. doi: 10.1038/emboj.2011.349. Epub 2011 Sep 27. EMBO J. 2012. PMID: 21952045 Free PMC article. - Yeast Pif1 accelerates annealing of complementary DNA strands.
Ramanagoudr-Bhojappa R, Byrd AK, Dahl C, Raney KD. Ramanagoudr-Bhojappa R, et al. Biochemistry. 2014 Dec 9;53(48):7659-69. doi: 10.1021/bi500746v. Epub 2014 Nov 26. Biochemistry. 2014. PMID: 25393406 Free PMC article. - Suppression of chromosome healing and anticheckpoint pathways in yeast postsenescence survivors.
Lai X, Heierhorst J. Lai X, et al. Genetics. 2013 Jun;194(2):403-8. doi: 10.1534/genetics.113.150813. Epub 2013 Mar 27. Genetics. 2013. PMID: 23535383 Free PMC article.
References
- Harper JW, Elledge SJ. Mol Cell. 2007;28:739–745. - PubMed
- Myung K, Datta A, Kolodner RD. Cell. 2001;104:397–408. - PubMed
- Diede SJ, Gottschling DE. Cell. 1999;99:723–733. - PubMed
- Marcand S, Brevet V, Mann C, Gilson E. Curr Biol. 2000;10:487–490. - PubMed
- Myung K, Chen C, Kolodner RD. Nature. 2001;411:1073–1076. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM26259/GM/NIGMS NIH HHS/United States
- R01 GM026259-31/GM/NIGMS NIH HHS/United States
- R01 GM026259/GM/NIGMS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R37 GM026259/GM/NIGMS NIH HHS/United States
- 84637/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous