A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein - PubMed (original) (raw)
. 2009 Nov;16(11):1134-40.
doi: 10.1038/nsmb.1680. Epub 2009 Oct 18.
Affiliations
- PMID: 19838190
- PMCID: PMC2784181
- DOI: 10.1038/nsmb.1680
A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein
Wataru Kamitani et al. Nat Struct Mol Biol. 2009 Nov.
Abstract
Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression, including type I interferon production, by promoting host mRNA degradation and inhibiting host translation, in infected cells. We present evidence that nsp1 uses a novel, two-pronged strategy to inhibit host translation and gene expression. Nsp1 bound to the 40S ribosomal subunit and inactivated the translational activity of the 40S subunits. Furthermore, the nsp1-40S ribosome complex induced the modification of the 5' region of capped mRNA template and rendered the template RNA translationally incompetent. Nsp1 also induced RNA cleavage in templates carrying the internal ribosome entry site (IRES) from encephalomyocarditis virus, but not in those carrying IRES elements from hepatitis C or cricket paralysis viruses, demonstrating that the nsp1-induced RNA modification was template-dependent. We speculate that the mRNAs that underwent the nsp1-mediated modification are marked for rapid turnover by the host RNA degradation machinery.
Figures
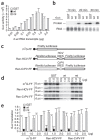
Figure 1. Effects of nsp1 on cap-dependent translation and IRES-mediated translation in RRL.
(a) Increasing amounts of rLuc RNA were incubated in RRL in the presence of 1 μg of nsp1 (wt), GST or nsp1-mt (mt) at 4 °C for 30 min; the molar ratios of rLuc RNA to nsp1 protein ranged from 1:10 to 1:2,000. Then the samples were incubated for 30 min at 30 °C, and the rLuc enzymatic activities were measured. y axis, light units. (b) A mixture of 0.25 μg of rLuc RNA and 1 μg of nsp1, GST or nsp1-mt was incubated in RRL in the presence of [35S]methionine at 30 °C for 10, 20 and 30 min. Translated products were analyzed on SDS-PAGE and detected by autoradiography. (c) Schematic diagram of various RNA transcripts used for d,e. (d) Increasing amounts of GST, nsp1, or nsp1-mt were incubated with RRL for 10 min at 4 °C. Then, the samples were incubated at 30 °C for 30 min with 0.5 μg of m7G-FF RNA, Ren-HCV-FF RNA, or Ren-CrPV-FF RNA in the presence of [35S]methionine. Translated products were analyzed on SDS-PAGE and detected by autoradiography. (e) For a given RNA group, the amount of fLuc protein, determined by densitometry, in each experimental group was normalized to the amount of the fLuc protein in a control group, to which the purified protein was not added. For a,d, averages of three independent experiments; error bars, s.d.
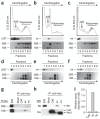
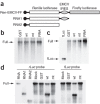
Figure 2. Binding of nsp1 to the 40S ribosomal subunits.
(a–c) CAT (a), nsp1 (b) or nsp1-mt (c) RNAs were transfected into 293 cells. Cell extracts were prepared 8 h after transfection and subjected to polysome profile analysis (top panels). CAT, nsp1 (wt) and nsp1-mt (mt) in each fraction were detected by western blot analysis using antibody to myc (middle panels). Bottom panels show rRNAs in each fraction. (d–f) After incubation of the mixture of 0.25 μg of rLuc RNA and 1 μg of nsp1, GST or nsp1-mt in RRL for 30 min at 30 °C, the samples were separated on 10%–50% sucrose gradient. GST, nsp1 (wt) and nsp1-mt (mt) in each fraction were detected by western blot analysis using anti-GST antibody and anti-nsp1 (ref. 8) antibody. The locations of GST (d), nsp1 (e) or nsp1-mt (f) in fractions are shown (all top panels). The bottom panels show rRNAs. (g–i) 293 cells were transfected with CAT RNA (CAT), nsp1 RNA (wt), or nsp1-mt RNA (mt). Cell extracts were prepared 8 h after transfection and subjected to immunoprecipitation using anti-myc antibody. Extracted RNAs from the immunoprecipitates were separated by agarose gel electrophoresis, and rRNAs were detected by northern blot analysis (g). The immunoprecipitated proteins were examined by western blot analysis using anti-S6 antibody (40S subunit specific) and anti-His antibody for CAT, nsp1 and nsp1-mt (h). Panel I represents the values, each of which was obtained by dividing the abundance of the immunoprecipitated 18S rRNA with the immunoprecipitated CAT protein, nsp1 protein or nsp1-mt protein in an arbitrary scale.
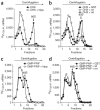
Figure 3. Ribosome binding assays of radiolabeled rLuc RNA.
Representative data from three independent experiments. (a) rLuc RNA was incubated in RRL in the presence of CHX only or a mixture of CHX and hippuristanol (hipp). The percentages of the rLuc RNA bound in 80S complexes were, for CHX, 17.4%; CHX + hipp., 1.0%. (b) rLuc RNA was incubated in RRL with GST, nsp1 (wt) or nsp1-mt (mt) in the presence of CHX. The percentages of the rLuc RNA bound in 80S complexes were, for GST, 13.3%; nsp1, 5.4%; nsp1-mt, 14.8%. (c) rLuc RNA was incubated in RRL in the presence of GMP-PNP or a mixture of GMP-PNP and hipp. The percentages of the rLuc RNA bound in 48S complexes were, for GMP-PNP, 11.7%; GMP-PNP + hipp., 2.9%. (d) rLuc RNA was incubated in RRL with GST, nsp1 (wt) or nsp1-mt (mt), in the presence of GMP-PNP. The percentages of the rLuc RNA bound in 48S complexes were, for GST, 13.1%; nsp1, 12.1%; nsp1-mt, 12.0%.
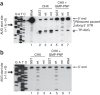
Figure 4. Toeprinting and primer extension analyses.
(a) Toeprinting. Samples shown in lanes 2–4 and 5–7 were incubated with CHX and a mixture of CHX and GMP-PNP, respectively, and toeprinting analysis was performed in the presence of GST (lanes 2 and 5), nsp1 (lanes 3 and 6) or nsp1-mt (lanes 4 and 7). Lane 1, the primer extension products using the rLuc RNA that was hybridized with a 5′-end labeled primer; 5′ end, the primer extension product that was extended to the 5′ end of rLuc RNA; UTR, untranslated region; TP-AUG, a correctly positioned toeprint. Arrowhead and asterisk represent a signal generated by the joining of the 60S subunits to the 40S complex positioned at the AUG codon. A dideoxynucleotide sequence of the rLuc gene generated with the same primer was run in parallel (left four rows).Translation initiation codon is shown at the left side of the gel. (b) Primer extension analysis. RRL was incubated with GST (lanes 1 and 4), nsp1 (lanes 2 and 5), or nsp1-mt (lanes 3 and 6) in the presence of CHX (lanes 1–3) or a mixture of CHX and GMP-PNP (lanes 4–6) for 5 min at 30 °C. Then rLuc RNA was added to each sample, and the samples were incubated for another 10 min at 30 °C. The RNAs were extracted and subjected to primer extension analysis. Lane 7, the primer extension products using the rLuc RNA that was hybridized with a 5′-end labeled primer.
Figure 5. Nsp1-induced modification of rLuc RNA in RRL.
(a) Cap-labeled rLuc RNA and the 3′-end-labeled rLuc RNA were incubated with GST, nsp1 (wt) or nsp1-mt (mt) in RRL for 15 min at 30 °C in the presence of CHX and GMP-PNP. RNAs were extracted, resolved on a sequencing gel and visualized by autoradiography. Asterisk, a signal that migrated slightly faster than the full-length rLuc in the sample incubated with nsp1. The 28S and 18S rRNAs, detected by ethidium bromide staining, are shown as loading controls. (b) Densitometric determination of rLuc RNAband intensities in a, normalized to the value of the GST-incubated sample. For the 3′-labeled rLuc RNA in the nsp1 sample, graph shows a sum of the signal intensities of the full-length rLuc RNA band and the band marked by the asterisk in a. (c) RRL was incubated with GST, nsp1 (wt), or nsp1-mt (mt) for 10 min at 4 °C. Then rLuc RNA was added and samples incubated at 30 °C for 20 min in the presence of [35S]methionine (1st translation). Aliquots were analyzed by SDS-PAGE. After addition of the same amount of CAT RNA to each sample, RNAs were extracted and translated using fresh RRL in the presence of [35S]methionine (2nd translation), and the samples were analyzed on a 12.5% SDS-PAGE gel. (d) Quantitation of rLuc protein abundances in the 1st and 2nd translation reactions in experiment in c. For b,d, averages of three independent experiments; error bars, s.d.
Figure 6. Nsp1 induces cleavage at the IRES in Ren-EMCV-FF RNA.
(a) Schematic diagram of Ren-EMCV-FF RNA and control RNAs RNA1 and RNA2. (b,c) After incubation of RRL with GST, nsp1 (wt), or nsp1-mt (mt) for 10 min at 4 °C, Ren-HCV-FF RNA (b) or Ren-CrPV-FF RNA (c) was added to each sample and incubated at 30 °C for 25 min. Then RNAs were extracted and analyzed by northern blot using a digoxigenin-labeled probe that binds to the fLuc ORF. RNA lane in b, the untreated Ren-HCV-FF RNA; fLuc lane in c, the RNA encoding fLuc ORF; RNA, the untreated Ren-CrPV-FF RNA. (d) RRL was incubated with GST, nsp1 (wt), or nsp1-mt (mt) for 10 min at 4 °C. Ren-EMCV-FF RNA was added to each sample and incubated at 30 °C for 10 min. RNAs were extracted and analyzed by northern blot using a probe that binds to the rLuc ORF (rLuc probe) or fLuc ORF (fLuc probe). Samples in the first three lanes, including untreated Ren-EMCV-FF (RNA), RNA1 and RNA2, served as size markers.
Figure 7. Importance of ribosomes for the function of nsp1.
(a) Cap-labeled rLuc RNA was incubated with GST, nsp1 (wt), or nsp1-mt (mt) in ribosome-free RRL for 15 min at 30 °C in the presence of CHX and GMP-PNP. RNAs were extracted, resolved on a sequencing gel and visualized by autoradiography. (b) The band intensity of rLuc RNA shown in a was determined by densitometric analysis. Mean of three independent experiments. (c) Experiments performed as described in Figure 5c, except that ribosome-free RRL was used for the initial translation reaction. (d) Cap-labeled rLuc RNA was incubated with GST, nsp1 (wt), or nsp1-mt (mt) in RRL in the presence of hippuristanol. RNAs were extracted, resolved on a sequencing gel and visualized by autoradiography.
Figure 8. Nsp1 inactivates the translational function of 40S ribosomal subunit.
RRL was incubated with GST, nsp1 (wt), or nsp1-mt (mt) for 10 min at 4 °C. Ren-CrPV-FF RNA was added to each sample and incubated at 30 °C for 25 min in the presence of [35S]methionine. Aliquots of the samples were used for SDS-PAGE (a, 1st). RNAs were extracted from the remaining samples after addition of the same amount of CAT RNA to each sample. Experiments were performed as described in Figure 5c, except that Ren-CrPV-FF was used as the template RNA. rLuc and fLuc are expressed by cap-dependent and CrPV IRES-mediated mechanisms, respectively (a, 2nd). (b) Experiments described in a were repeated three times, and the amounts of fLuc and rLuc proteins in the 2nd translation reaction are shown. Error bars, s.d. (c) Cap-labeled Ren-CrPV-FF RNA was incubated with GST, nsp1, or nsp1-mt in RRL for 15 min at 30 °C in the presence of CHX and GMP-PNP. RNAs were extracted, resolved on a sequencing gel and visualized by autoradiography.
Similar articles
- SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage.
Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. Huang C, et al. PLoS Pathog. 2011 Dec;7(12):e1002433. doi: 10.1371/journal.ppat.1002433. Epub 2011 Dec 8. PLoS Pathog. 2011. PMID: 22174690 Free PMC article. - Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation.
Lokugamage KG, Narayanan K, Huang C, Makino S. Lokugamage KG, et al. J Virol. 2012 Dec;86(24):13598-608. doi: 10.1128/JVI.01958-12. Epub 2012 Oct 3. J Virol. 2012. PMID: 23035226 Free PMC article. - Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA.
Tanaka T, Kamitani W, DeDiego ML, Enjuanes L, Matsuura Y. Tanaka T, et al. J Virol. 2012 Oct;86(20):11128-37. doi: 10.1128/JVI.01700-12. Epub 2012 Aug 1. J Virol. 2012. PMID: 22855488 Free PMC article. - Mechanisms of Coronavirus Nsp1-Mediated Control of Host and Viral Gene Expression.
Nakagawa K, Makino S. Nakagawa K, et al. Cells. 2021 Feb 2;10(2):300. doi: 10.3390/cells10020300. Cells. 2021. PMID: 33540583 Free PMC article. Review. - I(nsp1)ecting SARS-CoV-2-ribosome interactions.
Simeoni M, Cavinato T, Rodriguez D, Gatfield D. Simeoni M, et al. Commun Biol. 2021 Jun 10;4(1):715. doi: 10.1038/s42003-021-02265-0. Commun Biol. 2021. PMID: 34112887 Free PMC article. Review.
Cited by
- Role of IFN and Complements System: Innate Immunity in SARS-CoV-2.
Shibabaw T, Molla MD, Teferi B, Ayelign B. Shibabaw T, et al. J Inflamm Res. 2020 Sep 9;13:507-518. doi: 10.2147/JIR.S267280. eCollection 2020. J Inflamm Res. 2020. PMID: 32982366 Free PMC article. Review. - Emerging of a SARS-CoV-2 viral strain with a deletion in nsp1.
Benedetti F, Snyder GA, Giovanetti M, Angeletti S, Gallo RC, Ciccozzi M, Zella D. Benedetti F, et al. J Transl Med. 2020 Aug 31;18(1):329. doi: 10.1186/s12967-020-02507-5. J Transl Med. 2020. PMID: 32867854 Free PMC article. - The molecular virology of coronaviruses.
Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA. Hartenian E, et al. J Biol Chem. 2020 Sep 11;295(37):12910-12934. doi: 10.1074/jbc.REV120.013930. Epub 2020 Jul 13. J Biol Chem. 2020. PMID: 32661197 Free PMC article. Review. - Coronavirus cis-Acting RNA Elements.
Madhugiri R, Fricke M, Marz M, Ziebuhr J. Madhugiri R, et al. Adv Virus Res. 2016;96:127-163. doi: 10.1016/bs.aivir.2016.08.007. Epub 2016 Sep 6. Adv Virus Res. 2016. PMID: 27712622 Free PMC article. Review. - Viral and Cellular mRNA Translation in Coronavirus-Infected Cells.
Nakagawa K, Lokugamage KG, Makino S. Nakagawa K, et al. Adv Virus Res. 2016;96:165-192. doi: 10.1016/bs.aivir.2016.08.001. Epub 2016 Sep 10. Adv Virus Res. 2016. PMID: 27712623 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI072493/AI/NIAID NIH HHS/United States
- R01 AI072493-01A1/AI/NIAID NIH HHS/United States
- R01 AI072493-02/AI/NIAID NIH HHS/United States
- AI72493/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous