PIKfyve-ArPIKfyve-Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis - PubMed (original) (raw)
PIKfyve-ArPIKfyve-Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis
Ognian C Ikonomov et al. J Biol Chem. 2009.
Abstract
The phosphatidylinositol 3,5-bisphosphate (PtdIns(3,5)P(2)) metabolizing enzymes, the kinase PIKfyve and the phosphatase Sac3, constitute a single multiprotein complex organized by the PIKfyve regulator ArPIKfyve and its ability to homodimerize. We previously established that PIKfyve is activated within the triple PIKfyve-ArPIKfyve-Sac3 (PAS) core. These data assign an atypical function for the phosphatase in PtdIns(3,5)P(2) biosynthesis, thus raising the question of whether Sac3 retains its PtdIns(3,5)P(2) hydrolyzing activity within the PAS complex. Herein, we address the issue of Sac3 functionality by a combination of biochemical and morphological assays in triple-transfected COS cells using a battery of truncated or point mutants of the three proteins. We identified the Cpn60_TCP1 domain of PIKfyve as a major determinant for associating the ArPIKfyve-Sac3 subcomplex. Neither Sac3 nor PIKfyve enzymatic activities affected the PAS complex formation or stability. Using the well established formation of aberrant cell vacuoles as a sensitive functional measure of localized PtdIns(3,5)P(2) reduction, we observed a mitigated vacuolar phenotype by kinase-deficient PIKfyve(K1831E) if its ArPIKfyve-Sac3 binding region was deleted, suggesting reduced Sac3 access to, and turnover of PtdIns(3,5)P(2). In contrast, PIKfyve(K1831E), which displays intact ArPIKfyve-Sac3 binding, triggered a more severe vacuolar phenotype if coexpressed with ArPIKfyve(WT)-Sac3(WT) but minimal defects when coexpressed with ArPIKfyve(WT) and phosphatase-deficient Sac3(D488A). These data indicate that Sac3 assembled in the PAS regulatory core complex is an active PtdIns(3,5)P(2) phosphatase. Based on these and other data, presented herein, we propose a model of domain interactions within the PAS core and their role in regulating the enzymatic activities.
Figures
FIGURE 1.
ArPIKfyve and Sac3 associate with each other through their C termini. A, Sac3 deletion and point mutants are presented schematically relative to the Sac domain structure, predicted by Swiss-Prot and Smart databases (E-value cutoff = 1). Shown is a quantitative summary of their interaction with ArPIKfyveWT based on coimmunoprecipitation in COS7 cells, exemplified in B. B, cells cotransfected with cDNAs of Myc-ArPIKfyveWT and either eGFP-Sac3WT, eGFP-Sac3-(1–315), eGFP-Sac3-(1–574), eGFP-Sac3-(388–907), eGFP-Sac3-(478–907), eGFP-Sac3-(610–907), or eGFP-Sac3D488A were analyzed by immunoprecipitation (IP) with anti-Myc or anti-GFP antibodies as indicated. C, ArPIKfyve deletion mutants are presented schematically relative to the ArPIKfyve domain architecture predicted by Pfam databases (E-value cutoff = 1) and COILS. Shown is quantitation of their interaction with Sac3WT without or with differently tagged ArPIKfyveWT and measured by co-IP as shown in D and E. D, cells cotransfected with cDNAs of Myc-Sac3WT plus either eGFP-HA-ArPIKfyveWT, eGFP-HA- ArPIKfyveNt, or eGFP-ArPIKfyveCt were analyzed by IP with anti-Myc antibody are shown. E, cells double-transfected with cDNAs of Myc-ArPIKfyveWT plus eGFP-HA-ArPIKfyveWT (a control for ArPIKfyve homodimerization) or triple-transfected with cDNAs of Myc-ArPIKfyveWT, Myc-Sac3WT, and either eGFP-HA-ArPIKfyveWT or eGFP-HA- ArPIKfyveNt, were analyzed by IP with anti-HA antibodies. Seen is efficient co-IP of Myc-Sac3WT with the Myc-ArPIKfyveWT-eGFP-HA-ArPIKfyveWT homodimers. B, D, and E, fresh RIPA+ lysates were subjected to IP, and after washes in the same buffer, IPs were analyzed by SDS-PAGE and immunoblotting with anti-GFP and anti-Myc antibodies with a stripping step in between. Shown are chemiluminescence detections of a representative experiment for each of B, D, and E panels from three to six independent determinations with S.E. <10% of the mean value, quantitatively scored in A and C. Levels of the co-IP proteins were quantified relative to their total expression amounts and then normalized for the immunoprecipitated levels relative to the wild-type, which was scored as ++++. Mutants scored as − displayed <10% of the wild-type association.
FIGURE 2.
ArPIKfyve and Sac3 N-terminal regions interact with PIKfyve. A, in vitro associations of purified recombinant GST-PIKfyve, His6-ArPIKfyve, and Sac3. GST-PIKfyve or GST control purified from infected Sf9 cells and immobilized on GSH-agarose beads was incubated with bacterially produced His6-ArPIKfyve or His6-GDI2 (as a control), both purified on Ni-NTA-agarose (left panels), or with His6-ArPIKfyve-Sac3 subcomplex, purified on Ni-NTA-agarose from infected Sf21 cells (right panels) as indicated. Blots, cut in the middle, were probed with anti-PIKfyve (upper halves) or anti-ArPIKfyve and anti-Sac3 antibodies (lower halves) with a stripping step between the latter two. Shown are representative blots illustrating that efficient associations occur only with the three purified recombinant proteins. The asterisk (*) depicts incompletely stripped His6-ArPIKfyve. B, COS7 cells were triple-transfected with cDNAs encoding Myc-ArPIKfyveWT, HA-PIKfyveWT, and either eGFP-Sac3WT, eGFP-Sac3-(388–907), or eGFP-Sac3-(610–907). Fresh RIPA+ lysates underwent IP with anti-Myc antibody, and after washes in the same buffer, IPs were analyzed by SDS-PAGE and immunoblotting with anti-GFP, anti-Myc, and anti-HA-antibodies with a stripping step in between. Shown are chemiluminescence detections of a representative transfection experiment from three independent determinations with S.E. <10% of the mean. C, shown is a schematic diagram of the PIKfyve domain structure, predicted by Pfam (E-value cutoff = 1) or BLAST databases and a quantitative summary of the PIKfyve interaction with the complexes of ArPIKfyveWT-Sac3WT, ArPIKfyveWT-Sac3-(610–907), ArPIKfyveWT-Sac3(388–907), ArPIKfyve-(523–782)-Sac3WT, and ArPIKfyve-(298–782)-Sac3WT.
FIGURE 3.
PIKfyve regions associating with the ArPIKfyve-Sac3 complex. A, presented are the PIKfyve-truncated mutants and a quantitative summary of their interactions with ArPIKfyveWT-Sac3WT subcomplex measured by coimmunoprecipitation in COS7 cells (shown in B–G), the in vitro lipid kinase activity determined herein (see Fig. 4_A_ and
supplemental Fig. 2
) or in the indicated references, and their ability to induce vacuolar defects determined herein (see Fig. 4_B_ and data not shown) or in the indicated references. CHK homology (residues 1155–1461) harbors conserved Cys and His within its N-terminal half (residues 1155–1327) and conserved Lys within its C-terminal half (residues 1295–1461) that is homologous to spectrin repeats. B–G, COS7 cells were triple-transfected with Myc-Sac3WT, Myc-ArPIKfyveWT, and the following PIKfyve constructs: HA-PIKfyveWT, HA-PIKfyveΔCpn+, or eGFP-HA-PIKfyveΔFYVE (B); eGFP-HA-PIKfyveWT, eGFP-HA-PIKfyveΔC-Cpn, or eGFP-HA-PIKfyveΔN-Cpn (C); HA-PIKfyveWT or HA-PIKfyveΔSpecΔKin+ (D); eGFP-HA-PIKfyveWT or eGFP-HA-PIKfyveΔFΔKin+ (E); HA-PIKfyveWT, HA-PIKfyveK1831E, or HA-PIKfyveΔKin (F); eGFP-HA-PIKfyveWT or eGFP-HA-PIKfyveΔFΔDΔCpn (G). Anti-HA IPs from fresh RIPA+ lysates were analyzed by SDS-PAGE and immunoblotting with anti-HA, anti-Myc, and anti-GFP antibodies, with a stripping step in between. Shown are chemiluminescence detections from representative transfection experiments for each of the panels out of three to nine independent determinations quantified in A (mean ± S.E.). Levels of the co-IP proteins were quantified relative to their total expression amounts (seen in inputs) and then normalized for the immunoprecipitated levels of the indicated PIKfyve construct relative to PIKfyveWT, which was scored as 100%. A strong triple interaction was observed with PIKfyveWT, PIKfyveK1831E, and PIKfyveΔFYVE. The association is independent of whether PIKfyve carries GFP-HA or HA tags, but note the different electrophoretic mobility due to the different tag size.
FIGURE 4.
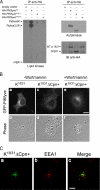
PIKfyveK1831E without the ArPIKfyve-Sac3 binding region is not dominantly interfering. A, fresh RIPA+ lysates, derived from COS cells singly transfected with cDNAs of HA-PIKfyveWT, HA-PIKfyveΔCpn+, HA-PIKfyveK1831E or with the empty vector underwent IP (in duplicate) with anti-HA antibodies. Washed IPs were subjected to assays for lipid kinase activity or autophosphorylation. Shown are autoradiograms of a plate with TLC-separated radiolabeled lipids and of the autokinase reaction resolved by SDS-PAGE and transferred onto a membrane, subsequently immunoblotted (IB) with anti-HA antibodies. Shown is a representative experiment of five to eight experiments with similar results. The lipid kinase activity is quantified by radioactive counting of the scraped silica and is shown in Fig. 3_A. B_, COS7 cells were transfected with cDNAs of eGFP-HA-PIKfyveK1831E or eGFP-HA-PIKfyveK1831EΔCpn+. Twenty hours post-transfection cells were treated with or without wortmannin (100 n
m
/20 min/37 °C, conditions that in COS cells do not induce wortmannin-dependent aberrant morphology; see Ref. 18) and then fixed in 4% formaldehyde. Fluorescence and phase-contrast images of transfected cells were captured by a Nikon Eclipse TE200 microscope (Apo 60×/1.40 Ph3DM oil objective) and processed as detailed under “Experimental Procedures.” Shown are typical images of the defective vacuolar phenotype induced by PIKfyveK1831E (panels a and b), a lack of vacuoles by HA-PIKfyveK1831EΔCpn+ seen in 94 ± 5% (mean ± S.E.) of transfected cells (panels c and d), and wortmannin sensitivity of the HA-PIKfyveK1831EΔCpn+ punctate staining observed in 88 ± 7% (mean ± S.E.) of treated transfected cells (panels e and f). C, cells were transfected with eGFP-HA-PIKfyveK1831EΔCpn+ cDNA and 24 h post-transfection processed for immunofluorescence microscopy (Olympus 1X81) with anti-EEA1. Confocal images (panels a and b) were processed by deconvolution analysis. The merge (panel c) of the two images indicates ∼40% colocalization of GFP-HA-PIKfyveK1831EΔCpn+ with EEA1. Bar, 10 μm.
FIGURE 5.
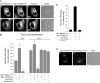
ArPIKfyveWT-Sac3WT exacerbates, whereas ArPIKfyveWT-Sac3D488A prevents the endomembrane defects by Sac3-binding PIKfyve mutants defective in PtdIns(3,5)P2 synthesis. A, COS7 cells were triple-cotransfected with cDNAs of eGFP-HA-PIKfyveK1831E, Myc-ArPIKfyveWT, and either HA-Sac3WT or HA-Sac3D488A. Twenty-four hours post-transfection cells were fixed in 4% formaldehyde and stained consecutively for Sac3 (anti-Sac3 IgG) followed by Alexa350 anti-rabbit secondary antibody) and ArPIKfyve (anti-Myc monoclonal antibody followed by Alexa568 anti-mouse secondary antibody). Fluorescence and phase-contrast images were captured by a Nikon Eclipse TE200 microscope (Apo 60×/1.40 Ph3DM oil objective) using three standard filter sets; green for eGFP, red for Alexa568, and blue for Alexa350. Note the presence of multiple vacuoles (panels a–d) under coexpression of enzymatically active Sac3WT and their absence under coexpression of inactive Sac3D488A (_panels e_-h), quantified in B. Bar, 10 μm. B, shown is quantification of the aberrant vacuolar phenotype in COS7 cells expressing pEGFP-HA-PIKfyveK1831E alone or in double and triple combinations with the indicated constructs. Cells showing clear translucent cytoplasmic vacuoles 12- and 24-h post-transfection were scored as positive. Data collected from examining >100 transfected cells per combination in three separate transfection experiments are presented as a percentage of the total number of inspected cells (mean ± S.E.; *, p < 0.01 or more versus eGFP-PIKfyveK1831E). In all double combinations with eGFP-HA-PIKfyveK1831E the aberrant vacuolar phenotype was seen to the same extent as in cells expressing eGFP-HA-PIKfyveK1831E alone. C, quantitation is shown of the vacuolar phenotype in COS7 cells expressing pEGFP-HA-PIKfyveK2000E alone or in a triple combination with ArPIKfyveWT-Sac3WT or ArPIKfyveWT-Sac3D488A scored by the presence of translucent cytoplasmic vacuoles 24-h post-transfection. Data were collected from examining at least 100 transfected cells per combination and are presented as a percentage of the total number of transfected cells (mean ± S.E.). Note that eGFP-HA-PIKfyveK2000E, partially deficient in PtdIns(3,5)P2 synthesis, was incapable of inducing vacuoles when expressed alone or with Sac3D488A-ArPIKfyveWT but readily triggered vacuolar defects when coexpressed with Sac3WT-ArPIKfyveWT. D, ArPIKfyveCt alleviates the phenotypic defects by PIKfyveK1831E. COS7 cells were transfected with cDNAs of Myc-PIKfyveK1831E and 3 h later with eGFP-ArPIKfyveCt. Twenty hours post-transfection cells were fixed and stained with anti-Myc monoclonal antibody and Alexa568 anti-mouse secondary antibody. Fluorescence (panels a and b) and phase-contrast images (panel c) were captured by a Nikon Eclipse TE200 microscope (Apo 60×/1.40 Ph3DM oil objective). Coexpressed ArPIKfyveCt diminished the number of PIKfyveK1831E-vacuolated cells by 42 ± 4%; p < 0.001 (based on observing 250 double-transfected cells in 2 separate experiments with consecutive transfections). Bar, 10 μm.
FIGURE 6.
Model for the interacting domains of the three proteins and regulated enzymatic activities in the PAS complex. PIKfyve associates with PtdIns(3)P-enriched endosome membranes via its FYVE finger, which is independent of the ArPIKfyve-Sac3 subcomplex. Without bound ArPIKfyve-Sac3, PIKfyve relays submaximal activity due to kinase-unfavorable conformation. The ArPIKfyve_n_-Sac3 complex, formed through an association of the C termini of an ArPIKfyve dimer (or higher-order homooligomer; hence ArPIKfyve_n_, where n ≥ 2) and the C terminus of a Sac3 monomer, docks to the Cpn60_TCP1 and the CH homology domains of PIKfyve. This induces conformational changes in PIKfyve to uncover binding sites at the catalytic domain, which associate with the N termini of ArPIKfyve_n_ and Sac3. These interactions stabilize a productive PAS core and allow PIKfyve to acquire an “activated” conformation. This transiently increases local synthesis of PtdIns(3,5)P2 that serves as recognition sites for PtdIns(3,5)P2 effectors. The local increase in PtdIns(3,5)P2 is counterbalanced by Sac3, which when incorporated in the PAS core, retains its activity for PtdIns(3,5)P2 hydrolysis. Neither the Sac3 nor the ArPIKfyve N terminus is dispensable for a productive PAS core complex even though a stable subcomplex via their C termini is formed. The PAS complex could also be formed in the cytosol and subsequently recruited to PtdIns(3)P on membranes.
Similar articles
- ArPIKfyve regulates Sac3 protein abundance and turnover: disruption of the mechanism by Sac3I41T mutation causing Charcot-Marie-Tooth 4J disorder.
Ikonomov OC, Sbrissa D, Fligger J, Delvecchio K, Shisheva A. Ikonomov OC, et al. J Biol Chem. 2010 Aug 27;285(35):26760-26764. doi: 10.1074/jbc.C110.154658. Epub 2010 Jul 14. J Biol Chem. 2010. PMID: 20630877 Free PMC article. - The PIKfyve-ArPIKfyve-Sac3 triad in human breast cancer: Functional link between elevated Sac3 phosphatase and enhanced proliferation of triple negative cell lines.
Ikonomov OC, Filios C, Sbrissa D, Chen X, Shisheva A. Ikonomov OC, et al. Biochem Biophys Res Commun. 2013 Oct 18;440(2):342-7. doi: 10.1016/j.bbrc.2013.09.080. Epub 2013 Sep 23. Biochem Biophys Res Commun. 2013. PMID: 24070605 Free PMC article. - Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex.
Sbrissa D, Ikonomov OC, Fu Z, Ijuin T, Gruenberg J, Takenawa T, Shisheva A. Sbrissa D, et al. J Biol Chem. 2007 Aug 17;282(33):23878-91. doi: 10.1074/jbc.M611678200. Epub 2007 Jun 7. J Biol Chem. 2007. PMID: 17556371 - PIKfyve and its Lipid products in health and in sickness.
Shisheva A. Shisheva A. Curr Top Microbiol Immunol. 2012;362:127-62. doi: 10.1007/978-94-007-5025-8_7. Curr Top Microbiol Immunol. 2012. PMID: 23086417 Review. - PIKfyve: Partners, significance, debates and paradoxes.
Shisheva A. Shisheva A. Cell Biol Int. 2008 Jun;32(6):591-604. doi: 10.1016/j.cellbi.2008.01.006. Epub 2008 Jan 25. Cell Biol Int. 2008. PMID: 18304842 Free PMC article. Review.
Cited by
- The determinants of head and neck cancer: Unmasking the PI3K pathway mutations.
Giudice FS, Squarize CH. Giudice FS, et al. J Carcinog Mutagen. 2013 Aug 2;Suppl 5:003. doi: 10.4172/2157-2518.S5-003. J Carcinog Mutagen. 2013. PMID: 25126449 Free PMC article. - Lipid kinases are essential for apicoplast homeostasis in Toxoplasma gondii.
Daher W, Morlon-Guyot J, Sheiner L, Lentini G, Berry L, Tawk L, Dubremetz JF, Wengelnik K, Striepen B, Lebrun M. Daher W, et al. Cell Microbiol. 2015 Apr;17(4):559-78. doi: 10.1111/cmi.12383. Epub 2014 Nov 22. Cell Microbiol. 2015. PMID: 25329540 Free PMC article. - Phosphoinositides and vesicular membrane traffic.
Mayinger P. Mayinger P. Biochim Biophys Acta. 2012 Aug;1821(8):1104-13. doi: 10.1016/j.bbalip.2012.01.002. Epub 2012 Jan 14. Biochim Biophys Acta. 2012. PMID: 22281700 Free PMC article. Review. - Insights into Lysosomal PI(3,5)P2 Homeostasis from a Structural-Biochemical Analysis of the PIKfyve Lipid Kinase Complex.
Lees JA, Li P, Kumar N, Weisman LS, Reinisch KM. Lees JA, et al. Mol Cell. 2020 Nov 19;80(4):736-743.e4. doi: 10.1016/j.molcel.2020.10.003. Epub 2020 Oct 23. Mol Cell. 2020. PMID: 33098764 Free PMC article. - PIP kinases: A versatile family that demands further therapeutic attention.
Llorente A, Arora GK, Grenier SF, Emerling BM. Llorente A, et al. Adv Biol Regul. 2023 Jan;87:100939. doi: 10.1016/j.jbior.2022.100939. Epub 2022 Dec 5. Adv Biol Regul. 2023. PMID: 36517396 Free PMC article.
References
- Balla T. (2006) J. Endocrinol. 188, 135–153 - PubMed
- Di Paolo G., De Camilli P. (2006) Nature 443, 651–657 - PubMed
- Poccia D., Larijani B. (2009) Biochem. J. 418, 233–246 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials