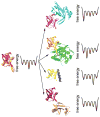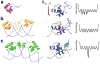The role of dynamic conformational ensembles in biomolecular recognition - PubMed (original) (raw)
Review
The role of dynamic conformational ensembles in biomolecular recognition
David D Boehr et al. Nat Chem Biol. 2009 Nov.
Erratum in
- Nat Chem Biol. 2009 Dec;5(12):954
Abstract
Molecular recognition is central to all biological processes. For the past 50 years, Koshland's 'induced fit' hypothesis has been the textbook explanation for molecular recognition events. However, recent experimental evidence supports an alternative mechanism. 'Conformational selection' postulates that all protein conformations pre-exist, and the ligand selects the most favored conformation. Following binding the ensemble undergoes a population shift, redistributing the conformational states. Both conformational selection and induced fit appear to play roles. Following binding by a primary conformational selection event, optimization of side chain and backbone interactions is likely to proceed by an induced fit mechanism. Conformational selection has been observed for protein-ligand, protein-protein, protein-DNA, protein-RNA and RNA-ligand interactions. These data support a new molecular recognition paradigm for processes as diverse as signaling, catalysis, gene regulation and protein aggregation in disease, which has the potential to significantly impact our views and strategies in drug design, biomolecular engineering and molecular evolution.
Figures
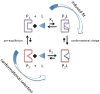
Figure 1
Thermodynamic cycle for molecular recognition processes involving induced fit or conformational selection. In conformational selection, the binding competent conformation (red, P2) is pre-existing in solution prior to the addition of ligand (L). The kinetic and thermodynamic rate constants can determine if conformational selection or induced fit is more likely,
Figure 2
Conformational selection in protein-ligand interactions observed by NMR R2 relaxation dispersion experiments. (a) Locations of conformational exchange are indicated as spheres on the structure of DHFR (pdb 1rx5). (b) A linear correlation between Δω (chemical shift difference between ground-state and higher energy conformations) from R2 relaxation dispersion experiments of the product binary complex of DHFR (enzyme bound with tetrahydrofolate (E:THF)) and Δδ from ground-state chemical shift differences between product binary and ternary (enzyme bound with tetrahydrofolate and NADPH cofactor (E:THF:NADPH)) complexes indicate that the higher energy conformation of the product binary complex is structurally similar to the ground-state of the product ternary complex (i.e. chemical shifts of the higher energy conformation of the product binary complex are similar to the chemical shifts of the ground-state conformation of the product ternary complex) (data taken from ref41). (c) The binding of the NADPH cofactor changes the free energy landscape of the enzyme. Structurally similar conformations are colored alike.
Figure 3
A schematic illustration of molecular recognition processes involving ubiquitin. The NMR-derived conformational ensemble of ubiquitin indicates that all bound conformations exist in the absence of protein binding partners (left). Although the conformational ensemble encompasses all forty six of the known crystal structures of ubiquitin, only five are shown here for clarity (pdb 1f9j, 1s1q, 1xd3, 2d36 and 2g45). The free energy landscapes are hypothetical considering that the relative population of each conformation in the ensemble and the energy barriers separating the conformations are not known.
Figure 4
DNA recognition by the lac repressor headpiece. Differences in structure (left) and dynamics (middle) between (A) lac repressor headpiece in the free state (PDB code 1lqc), (B) lac repressor headpiece bound to noncognate DNA (pdb 1osl) and (C) lac repressor headpiece bound to cognate DNA (1l1m). The middle column shows Rex, the contribution to the NMR R2 transverse rate constant from μs-ms time scale conformational fluctuations, mapped onto the structures of the lac repressor protein. (Adapted from ref with permission from the American Chemical Society). The free energy landscape (shown schematically on the right) is rough, with many interconverting substates in the complex of lac repressor headpiece with noncognate DNA, but a single dominant conformation is formed in the complex with cognate DNA.
Similar articles
- Understanding biomolecular motion, recognition, and allostery by use of conformational ensembles.
Fenwick RB, Esteban-Martín S, Salvatella X. Fenwick RB, et al. Eur Biophys J. 2011 Dec;40(12):1339-55. doi: 10.1007/s00249-011-0754-8. Epub 2011 Nov 17. Eur Biophys J. 2011. PMID: 22089251 Free PMC article. Review. - Evidence of conformational selection driving the formation of ligand binding sites in protein-protein interfaces.
Bohnuud T, Kozakov D, Vajda S. Bohnuud T, et al. PLoS Comput Biol. 2014 Oct 2;10(10):e1003872. doi: 10.1371/journal.pcbi.1003872. eCollection 2014 Oct. PLoS Comput Biol. 2014. PMID: 25275445 Free PMC article. - Single molecule insights on conformational selection and induced fit mechanism.
Hatzakis NS. Hatzakis NS. Biophys Chem. 2014 Feb;186:46-54. doi: 10.1016/j.bpc.2013.11.003. Epub 2013 Nov 13. Biophys Chem. 2014. PMID: 24342874 Review. - Pioneer in Molecular Biology: Conformational Ensembles in Molecular Recognition, Allostery, and Cell Function.
Nussinov R. Nussinov R. J Mol Biol. 2025 Jun 1;437(11):169044. doi: 10.1016/j.jmb.2025.169044. Epub 2025 Feb 25. J Mol Biol. 2025. PMID: 40015370 Review. - Conformational selection is a dominant mechanism of ligand binding.
Vogt AD, Di Cera E. Vogt AD, et al. Biochemistry. 2013 Aug 27;52(34):5723-9. doi: 10.1021/bi400929b. Epub 2013 Aug 15. Biochemistry. 2013. PMID: 23947609 Free PMC article.
Cited by
- The catalytic roles of P185 and T188 and substrate-binding loop flexibility in 3α-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni.
Hwang CC, Chang YH, Lee HJ, Wang TP, Su YM, Chen HW, Liang PH. Hwang CC, et al. PLoS One. 2013 May 23;8(5):e63594. doi: 10.1371/journal.pone.0063594. Print 2013. PLoS One. 2013. PMID: 23717450 Free PMC article. - Structural Architecture of Prothrombin in Solution Revealed by Single Molecule Spectroscopy.
Pozzi N, Bystranowska D, Zuo X, Di Cera E. Pozzi N, et al. J Biol Chem. 2016 Aug 26;291(35):18107-16. doi: 10.1074/jbc.M116.738310. Epub 2016 Jul 19. J Biol Chem. 2016. PMID: 27435675 Free PMC article. - Enhanced Conformational Sampling Using Replica Exchange with Collective-Variable Tempering.
Gil-Ley A, Bussi G. Gil-Ley A, et al. J Chem Theory Comput. 2015 Mar 10;11(3):1077-85. doi: 10.1021/ct5009087. J Chem Theory Comput. 2015. PMID: 25838811 Free PMC article. - Structure and mechanism of purine-binding riboswitches.
Batey RT. Batey RT. Q Rev Biophys. 2012 Aug;45(3):345-81. doi: 10.1017/S0033583512000078. Epub 2012 Jul 31. Q Rev Biophys. 2012. PMID: 22850604 Free PMC article. Review. - Unraveling the Coupling between Conformational Changes and Ligand Binding in Ribose Binding Protein Using Multiscale Molecular Dynamics and Free-Energy Calculations.
Ren W, Dokainish HM, Shinobu A, Oshima H, Sugita Y. Ren W, et al. J Phys Chem B. 2021 Mar 25;125(11):2898-2909. doi: 10.1021/acs.jpcb.0c11600. Epub 2021 Mar 17. J Phys Chem B. 2021. PMID: 33728914 Free PMC article.
References
- Fischer E. Einfluss der configuration auf die Wirkung der Enzyme. Ber Dtsch Chem Ges. 1894;27:2984–2993.
- Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–603. - PubMed
- Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA096865/CA/NCI NIH HHS/United States
- GM75995/GM/NIGMS NIH HHS/United States
- R01 GM075995/GM/NIGMS NIH HHS/United States
- N01 CO012400/CA/NCI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- N01 CO012400/CO/NCI NIH HHS/United States
- CA96865/CA/NCI NIH HHS/United States
- N01-CO-12400/CO/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous