Beta integrin tyrosine phosphorylation is a conserved mechanism for regulating talin-induced integrin activation - PubMed (original) (raw)
Beta integrin tyrosine phosphorylation is a conserved mechanism for regulating talin-induced integrin activation
Nicholas J Anthis et al. J Biol Chem. 2009.
Abstract
Integrins are large membrane-spanning receptors fundamental to cell adhesion and migration. Integrin adhesiveness for the extracellular matrix is activated by the cytoskeletal protein talin via direct binding of its phosphotyrosine-binding-like F3 domain to the cytoplasmic tail of the beta integrin subunit. The phosphotyrosine-binding domain of the signaling protein Dok1, on the other hand, has an inactivating effect on integrins, a phenomenon that is modulated by integrin tyrosine phosphorylation. Using full-length tyrosine-phosphorylated (15)N-labeled beta3, beta1A, and beta7 integrin tails and an NMR-based protein-protein interaction assay, we show that talin1 binds to the NPXY motif and the membrane-proximal portion of beta3, beta1A, and beta7 tails, and that the affinity of this interaction is decreased by integrin tyrosine phosphorylation. Dok1 only interacts weakly with unphosphorylated tails, but its affinity is greatly increased by integrin tyrosine phosphorylation. The Dok1 interaction remains restricted to the integrin NPXY region, thus phosphorylation inhibits integrin activation by increasing the affinity of beta integrin tails for a talin competitor that does not form activating membrane-proximal interactions with the integrin. Key residues governing these specificities were identified by detailed structural analysis, and talin1 was engineered to bind preferentially to phosphorylated integrins by introducing the mutation D372R. As predicted, this mutation affects talin1 localization in live cells in an integrin phosphorylation-specific manner. Together, these results indicate that tyrosine phosphorylation is a common mechanism for regulating integrin activation, despite subtle differences in how these integrins interact with their binding proteins.
Figures
FIGURE 1.
Integrin tail and PTB domain sequence alignments. A, sequences of the cytoplasmic regions of the β3, β1A, and β7 integrin tails. The two NP_X_Y motif tyrosine residues are indicated, with β3 numbering. The MP, nMD, and fMD regions are denoted. Secondary structure is based on the β1D·talin2 complex structure (44), with α helices denoted in blue and 310 helices in green. B, sequences of the PTB domains of Dok1, talin1, and talin2 aligned by secondary structure, with secondary structure elements from the Dok1 PTB domain structure (PDB 2V76) (43) shown. Notable residues are indicated with Dok1 numbering.
FIGURE 2.
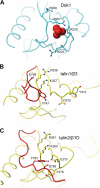
The structural basis of Dok1 specificity for phosphorylated integrin tails. A, detail of the Dok1 PTB domain structure (PDB 2V76) (43) showing a sulfate anion located in the NP_X_Y-binding pocket. Key positively charged residues are highlighted. B, detail of the NP_X_Y motif of the β3 integrin tail bound to the talin1 F3 domain (PDB 1MK9) (55). C, detail of the NP_X_Y motif of the β1D tail bound to the talin2 F3 domain (PDB 3G9W) (44). The residues highlighted in panels B and C are analogous to those highlighted in the Dok1 structure in panel A. Molecular images were generated with MOLMOL (61).
FIGURE 3.
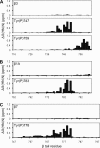
Phosphorylation of integrin tails. HSQC spectra of integrin tails before (red) and after (blue) the tyrosine phosphorylation reaction. Phosphorylated residues are indicated. A, β3 Y759F. B, β3 Y747F. C, β1A Y795F. D, β1A Y783F. E, β1A WT. F, β7 Y753F/Y758F.
FIGURE 4.
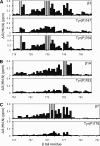
Effect of tyrosine phosphorylation on the integrin/talin1 interaction. Weighted chemical shift maps of perturbations observed in 1H-15N HSQC spectra of the β3 (A), β1A (B), and β7 (C) tails (50 μ
m
) upon the addition of talin1 F3 domain (1 m
m
). Interaction studies were performed on unphosphorylated integrin tails and tails phosphorylated at the nMD site (β3, β1A, and β7) and at the fMD site (β3). Gray bars correspond to residues that could not be tracked due to exchange broadening. Note that the y axis scale differs between panels.
FIGURE 5.
Effect of tyrosine phosphorylation on the integrin/Dok1 interaction. Weighted chemical shift maps of perturbations observed in 1H-15N HSQC spectra of the β3 (A), β1A (B), and β7 (C) tails (50 μ
m
) upon the addition of the Dok1 PTB domain (1 m
m
). Interaction studies were performed on unphosphorylated integrin tails and tails phosphorylated at the nMD site (β3, β1A, and β7) and at the fMD site (β3).
FIGURE 6.
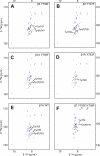
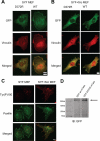
Talin D372R preferentially localizes to focal adhesions that are tyrosine phosphorylated. A, SYF MEFs and B, SYF + Src MEFs transiently expressing GFP-talin1 WT or D372R were allowed to adhere to fibronectin-coated coverslips and stained to visualize vinculin. Depicted are the localization of talin (green) and vinculin (red). C, SYF MEFs and SYF + Src MEFs were stained to visualize phosphotyrosine (Tyr(P)100, red) and paxillin (green). D, SYF cells expressing GFP-talin1 and GFP-talin1 D372R were lysed and analyzed by Western blotting to confirm comparable expression of D372R and WT talin1.
Similar articles
- An integrin phosphorylation switch: the effect of beta3 integrin tail phosphorylation on Dok1 and talin binding.
Oxley CL, Anthis NJ, Lowe ED, Vakonakis I, Campbell ID, Wegener KL. Oxley CL, et al. J Biol Chem. 2008 Feb 29;283(9):5420-6. doi: 10.1074/jbc.M709435200. Epub 2007 Dec 21. J Biol Chem. 2008. PMID: 18156175 - Interaction Analyses of 14-3-3ζ, Dok1, and Phosphorylated Integrin β Cytoplasmic Tails Reveal a Bi-molecular Switch in Integrin Regulation.
Chatterjee D, D'Souza A, Zhang Y, Bin W, Tan SM, Bhattacharjya S. Chatterjee D, et al. J Mol Biol. 2018 Oct 19;430(21):4419-4430. doi: 10.1016/j.jmb.2018.09.008. Epub 2018 Sep 20. J Mol Biol. 2018. PMID: 30243836 - An Alternative Phosphorylation Switch in Integrin β2 (CD18) Tail for Dok1 Binding.
Gupta S, Chit JC, Feng C, Bhunia A, Tan SM, Bhattacharjya S. Gupta S, et al. Sci Rep. 2015 Jun 25;5:11630. doi: 10.1038/srep11630. Sci Rep. 2015. PMID: 26108885 Free PMC article. - The talin-tail interaction places integrin activation on FERM ground.
Campbell ID, Ginsberg MH. Campbell ID, et al. Trends Biochem Sci. 2004 Aug;29(8):429-35. doi: 10.1016/j.tibs.2004.06.005. Trends Biochem Sci. 2004. PMID: 15362227 Review. - Talin controls integrin activation.
Calderwood DA. Calderwood DA. Biochem Soc Trans. 2004 Jun;32(Pt3):434-7. doi: 10.1042/BST0320434. Biochem Soc Trans. 2004. PMID: 15157154 Review.
Cited by
- Acetyl-NPKY of integrin-β1 binds KINDLIN2 to control endothelial cell proliferation and junctional integrity.
Sidibé A, Mykuliak VV, Zhang P, Hytönen VP, Wu J, Wehrle-Haller B. Sidibé A, et al. iScience. 2024 May 28;27(6):110129. doi: 10.1016/j.isci.2024.110129. eCollection 2024 Jun 21. iScience. 2024. PMID: 38904068 Free PMC article. - LFA-1 Activation in T-Cell Migration and Immunological Synapse Formation.
Shi H, Shao B. Shi H, et al. Cells. 2023 Apr 12;12(8):1136. doi: 10.3390/cells12081136. Cells. 2023. PMID: 37190045 Free PMC article. Review. - The structural basis of β2 integrin intra-cellular multi-protein complexes.
Bhattacharjya S. Bhattacharjya S. Biophys Rev. 2022 Sep 7;14(5):1183-1195. doi: 10.1007/s12551-022-00995-x. eCollection 2022 Oct. Biophys Rev. 2022. PMID: 36345283 Free PMC article. Review. - c-Met-integrin cooperation: Mechanisms, tumorigenic effects, and therapeutic relevance.
Stanislovas J, Kermorgant S. Stanislovas J, et al. Front Cell Dev Biol. 2022 Oct 14;10:994528. doi: 10.3389/fcell.2022.994528. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36330337 Free PMC article. Review. - High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Zeng J, Schmitz F, Isaksson S, Glas J, Arbab O, Andersson M, Sundell K, Eriksson LA, Swaminathan K, Törnroth-Horsefield S, Hedfalk K. Zeng J, et al. Life Sci Alliance. 2022 Oct 13;5(12):e202201491. doi: 10.26508/lsa.202201491. Life Sci Alliance. 2022. PMID: 36229063 Free PMC article.
References
- Hynes R. O. (2002) Cell 110, 673–687 - PubMed
- Liu S., Calderwood D. A., Ginsberg M. H. (2000) J. Cell Sci. 113, 3563–3571 - PubMed
- Ginsberg M. H., Partridge A., Shattil S. J. (2005) Curr. Opin. Cell Biol. 17, 509–516 - PubMed
- Campbell I. D., Ginsberg M. H. (2004) Trends Biochem. Sci. 29, 429–435 - PubMed
- Calderwood D. A. (2004) J. Cell Sci. 117, 657–666 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous