Evidence that localized variation in primate sequence divergence arises from an influence of nucleosome placement on DNA repair - PubMed (original) (raw)
Evidence that localized variation in primate sequence divergence arises from an influence of nucleosome placement on DNA repair
Hua Ying et al. Mol Biol Evol. 2010 Mar.
Abstract
Understanding the origins of localized substitution rate heterogeneity has important implications for identifying functional genomic sequences. Outside of gene regions, the origins of rate heterogeneity remain unclear. Experimental studies establish that chromatin compaction affects rates of both DNA lesion formation and repair. A functional association between chromatin status and 5-methyl-cytosine also exists. These suggest that both the total rate and the type of substitution will be affected by chromatin status. Regular positioning of nucleosomes, the building block of chromatin, further predicts that substitution rate and type should vary spatially in an oscillating manner. We addressed chromatin's influence on substitution rate and type in primates. Matched numbers of sites were sampled from Dnase I hypersensitive (DHS) and closed chromatin control flank (Flank). Likelihood ratio tests revealed significant excesses of total and of transition substitutions in Flank compared with matched DHS for both intergenic and intronic samples. An additional excess of CpG transitions was evident for the intergenic, but not intronic, regions. Fluctuation in substitution rate along approximately 1,800 primate promoters was measured using phylogenetic footprinting. Significant positive correlations were evident between the substitution rate and a nucleosome score from resting human T-cells, with up to approximately 50% of the variance in substitution rate accounted for. Using signal processing techniques, a dominant oscillation at approximately 200 bp was evident in both the substitution rate and the nucleosome score. Our results support a role for differential DNA repair rates between open and closed chromatin in the spatial distribution of rate heterogeneity.
Figures
FIG. 1.
Comparison of the substitution signal estimated using phylogenetic footprinting and a phylo-HMM. Shown in the top row of panels is substitution rate variation from footprinting, measured as the sum of tree branch lengths (K), from the genes C D _X_2 and F G _F_5. The lower panel row shows the posterior probabilities a site was classified as fast (_p_fast), estimated from the phylo-HMM. Each horizontal line indicates a nucleosome inferred from one of seven cell lines where magenta represents one of the four cancer cell lines of A375, T47D, MCF7, and MALME; green represents IMR90 cell line; cyan represents PM cell line; and yellow represents the MEC cell line (Ozsolak et al. 2007).  is the estimated Pearson's correlation coefficient of the footprinting and phylo-HMM signals.
is the estimated Pearson's correlation coefficient of the footprinting and phylo-HMM signals.
FIG. 2.
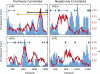
Comparison of the spatial substitution signal with nucleosome score. Example genes exhibiting a positive correlation are shown in the left column, a negative correlation in the right column. x axis is the alignment position; y axis black label is the nucleosome score (Schones et al. 2008) with data shown as a blue histogram; y axis red label is the estimate of K from footprinting with data shown as the red line. Black, orange, and blue horizontal lines with diamond marks at the end represent CpG islands, DHS sites, and repeat sequences, respectively.
FIG. 3.
Signal analysis of substitution amplitude spectra from DUSP and FZD2 promoters. The plot columns correspond to the indicated loci. The upper plot row shows K, whereas the lower row its DFT-based amplitude spectrum. Periods of the footprinting signal appear as peaks of the amplitude spectrum. The first, second, and third highest peaks are annotated with a corresponding number of +, and their period lengths, and CRB are shown in the tables.
FIG. 4.
Evolutionary distance and raw nucleosome score exhibit a ∼200-bp period in primate promoters. Frequency histograms of the periods classified as main (upper row) and secondary (lower row) after eliminating periods with a CRB > 0.2. The left and right columns show the periods from the substitution spectra (K) and the nucleosome score (Schones et al. 2008), respectively.
Similar articles
- Defining Regulatory Elements in the Human Genome Using Nucleosome Occupancy and Methylome Sequencing (NOMe-Seq).
Rhie SK, Schreiner S, Farnham PJ. Rhie SK, et al. Methods Mol Biol. 2018;1766:209-229. doi: 10.1007/978-1-4939-7768-0_12. Methods Mol Biol. 2018. PMID: 29605855 Free PMC article. - Contrasting chromatin organization of CpG islands and exons in the human genome.
Choi JK. Choi JK. Genome Biol. 2010;11(7):R70. doi: 10.1186/gb-2010-11-7-r70. Epub 2010 Jul 5. Genome Biol. 2010. PMID: 20602769 Free PMC article. - DNase-seq predicts regions of rotational nucleosome stability across diverse human cell types.
Winter DR, Song L, Mukherjee S, Furey TS, Crawford GE. Winter DR, et al. Genome Res. 2013 Jul;23(7):1118-29. doi: 10.1101/gr.150482.112. Epub 2013 May 8. Genome Res. 2013. PMID: 23657885 Free PMC article. - Chromatin 'programming' by sequence--is there more to the nucleosome code than %GC?
Hughes A, Rando OJ. Hughes A, et al. J Biol. 2009;8(11):96. doi: 10.1186/jbiol207. Epub 2009 Dec 23. J Biol. 2009. PMID: 20067596 Free PMC article. Review. - DNA methylation, nucleosome formation and positioning.
Pennings S, Allan J, Davey CS. Pennings S, et al. Brief Funct Genomic Proteomic. 2005 Feb;3(4):351-61. doi: 10.1093/bfgp/3.4.351. Brief Funct Genomic Proteomic. 2005. PMID: 15814025 Review.
Cited by
- Predicting regional somatic mutation rates using DNA motifs.
Liu C, Wang Z, Wang J, Liu C, Wang M, Ngo V, Wang W. Liu C, et al. PLoS Comput Biol. 2023 Oct 2;19(10):e1011536. doi: 10.1371/journal.pcbi.1011536. eCollection 2023 Oct. PLoS Comput Biol. 2023. PMID: 37782656 Free PMC article. - Environmental carcinogens disproportionally mutate genes implicated in neurodevelopmental disorders.
Baker BH, Zhang S, Simon JM, McLarnan SM, Chung WK, Pearson BL. Baker BH, et al. Front Neurosci. 2023 Aug 3;17:1106573. doi: 10.3389/fnins.2023.1106573. eCollection 2023. Front Neurosci. 2023. PMID: 37599994 Free PMC article. - Chromatin structure influences rate and spectrum of spontaneous mutations in Neurospora crassa.
Villalba de la Peña M, Summanen PAM, Liukkonen M, Kronholm I. Villalba de la Peña M, et al. Genome Res. 2023 Apr;33(4):599-611. doi: 10.1101/gr.276992.122. Epub 2023 Mar 15. Genome Res. 2023. PMID: 36922001 Free PMC article. - Non-B DNA: a major contributor to small- and large-scale variation in nucleotide substitution frequencies across the genome.
Guiblet WM, Cremona MA, Harris RS, Chen D, Eckert KA, Chiaromonte F, Huang YF, Makova KD. Guiblet WM, et al. Nucleic Acids Res. 2021 Feb 22;49(3):1497-1516. doi: 10.1093/nar/gkaa1269. Nucleic Acids Res. 2021. PMID: 33450015 Free PMC article. - [Identification of nucleosome positioning using support vector machine method based on comprehensive DNA sequence feature].
Cui Y, Xu Z, Li J. Cui Y, et al. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2020 Jun 25;37(3):496-501. doi: 10.7507/1001-5515.201911064. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2020. PMID: 32597092 Free PMC article. Chinese.
References
- Anastassiou D. Genomic signal processing. IEEE Signal Process Mag. 2001;18(4):8–20.
- Balhorn R, Weston S, Thomas C, Wyrobek AJ. DNA packaging in mouse spermatids. Synthesis of protamine variants and four transition proteins. Exp Cell Res. 1984;150(2):298–308. - PubMed
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40(2):359–369. - PubMed
- Boulikas T. Evolutionary consequences of nonrandom damage and repair of chromatin domains. J Mol Evol. 1992;35(2):156–180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources