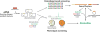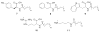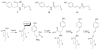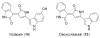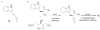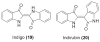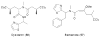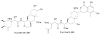Metagenomic approaches to natural products from free-living and symbiotic organisms - PubMed (original) (raw)
Review
. 2009 Nov;26(11):1488-503.
doi: 10.1039/b817078a. Epub 2009 Sep 16.
Affiliations
- PMID: 19844642
- PMCID: PMC2919151
- DOI: 10.1039/b817078a
Review
Metagenomic approaches to natural products from free-living and symbiotic organisms
Sean F Brady et al. Nat Prod Rep. 2009 Nov.
Abstract
Bacterial cultivation has been a mainstay of natural products discovery for the past 80 years. However, the majority of bacteria are recalcitrant to culture, providing an untapped source for new natural products. Metagenomic analysis provides an alternative method to directly access the uncultivated genome for natural products research and for the discovery of novel, bioactive substances. Applications of metagenomics to diverse habitats, such as soils and the interior of animals, are described.
Figures
Figure 1
Overview of culture independent strategies used for the discovery of natural products and natural product gene clusters from eDNA libraries.
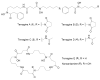
Figure 2
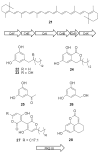
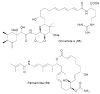
The terragines (1-5) and norcardamine (6) were characterized from S. lividans transformed with soil eDNA cosmids.
Figure 3
Families of N-acyl amino acid antibiotics characterized from eDNA clones. In each case a collection of metabolites with both saturated and monounsaturated side chains of various lengths are produced by the antibacterially active eDNA clone.
Figure 4
Long chain N-acyltyrosine antibiotics (7) have been characterized from a number of different antibacterially active eDNA clones. One of these clones was found to produce additional families of metabolites (8 and 9). In this clone the fee gene cluster is responsible for the biosynthesis of all three families of compounds.
Figure 5
The previously reported metabolites violacein (14) and deoxyviolacein (15) are produced by a purple eDNA cosmid clone.
Figure 6
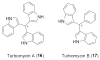
Turbomycin A and B are tri-aryl cations that accumulate in the acid precipitate of melanin producing eDNA clones at a higher level than is observed in acid precipitates from vector control cultures.
Figure 7
The isonitrile functionalized indole derivative 18 (a) is produced in E. coli from tryptophan and a five-carbon sugar using the eDNA-derived enzymes IsnA and IsnB (b).
Figure 8
Three different metagenomic studies have identified eDNA clones that produce indigo and indirubin.
Figure 9
Both known (21-26) and novel (27 and 28) small molecules were characterized from clones found in phenotypic screens of R. metallidurans based libraries.
Figure 10
KSβ sequences were PCR amplified from eDNA and these eDNA derived KSβ sequences were used to construct hybrid minimal PKSs. The resulting hybrid minimal PKS cassettes were then shuttled into Streptomyces and found to produce compounds 29-32.
Figure 11
Diene alcohols (33, 34) found during the analysis of culture broth extracts from cultures of S. lividans transformed with a soil eDNA cosmid clone (X+Y=12).
Figure 12
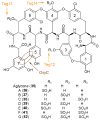
The VEG and TEG glycopeptide gene clusters were cloned from a soil eDNA mega library. Sulfo-teicoplanins A-G (36-42) were produced from the teicoplanin aglycone (35) using three sulfotransferases (Teg 13, 14, and 15) found in the eDNA-derived TEG gene cluster. Gene cluster color coding: nonribosomal peptide synthetase (green), amino acid biosynthesis (green hashed arrows), glycosyl transferase (red), sugar biosynthesis (red hashed arrows), oxidative coupling (brown), methyl transferase (blue), sulfotransferase (orange), halogenase (yellow).
Figure 13
Genes for the biosynthesis of two polyketides were more readily obtained because of guiding microbiology that simplified the problem.
Figure 14
Cyanobactin biosynthetic genes were cloned from enriched samples of Prochloron symbiotic bacteria living in ascidian animals.
Figure 15
Two metabolites originally isolated from fungi were later shown to originate in endosymbiotic bacteria.
Figure 16
Polyketides from ant symbionts (50) and, putatively, a sponge symbiont (51).
Figure 17
Compounds that helped with enabling technologies.
Figure 18
Some polyketides from sponges and ascidians.
Figure 19
Halogenated peptides, putatively from sponge symbionts.
Figure 20
Further probable _trans_-AT products from sponges.
Figure 21
Use of metagenomics in engineering. Top: The natural product ulithiacyclamide (46) was “converted” to the wholly unnatural compound eptidemnamide (61) by (a) one-step PCR mutagenesis and synthesized in E. coli. Bottom: The natural product patellin 2 was converted to the rare natural product trunkamide (47) by (b) one-step recombination in yeast, followed by production in E. coli. These results and the natural selectivity of the enzymes indicate that large libraries of products can be synthesized by identical enzymes.
Similar articles
- Using genomics to deliver natural products from symbiotic bacteria.
Clardy J. Clardy J. Genome Biol. 2005;6(9):232. doi: 10.1186/gb-2005-6-9-232. Epub 2005 Aug 31. Genome Biol. 2005. PMID: 16168093 Free PMC article. Review. - Origin and variation of tunicate secondary metabolites.
Schmidt EW, Donia MS, McIntosh JA, Fricke WF, Ravel J. Schmidt EW, et al. J Nat Prod. 2012 Feb 24;75(2):295-304. doi: 10.1021/np200665k. Epub 2012 Jan 10. J Nat Prod. 2012. PMID: 22233390 Free PMC article. Review. - Natural products genomics.
Siezen RJ, Khayatt BI. Siezen RJ, et al. Microb Biotechnol. 2008 Jul;1(4):275-82. doi: 10.1111/j.1751-7915.2008.00044.x. Microb Biotechnol. 2008. PMID: 21261848 Free PMC article. No abstract available. - Cellular targets of natural products.
Dixon N, Wong LS, Geerlings TH, Micklefield J. Dixon N, et al. Nat Prod Rep. 2007 Dec;24(6):1288-310. doi: 10.1039/b616808f. Epub 2007 Oct 17. Nat Prod Rep. 2007. PMID: 18033580 Review. - Ecological and biotechnological importance of secondary metabolites produced by coral-associated bacteria.
Modolon F, Barno AR, Villela HDM, Peixoto RS. Modolon F, et al. J Appl Microbiol. 2020 Dec;129(6):1441-1457. doi: 10.1111/jam.14766. Epub 2020 Jul 23. J Appl Microbiol. 2020. PMID: 32627318 Review.
Cited by
- Emerging strategies and integrated systems microbiology technologies for biodiscovery of marine bioactive compounds.
Rocha-Martin J, Harrington C, Dobson AD, O'Gara F. Rocha-Martin J, et al. Mar Drugs. 2014 Jun 10;12(6):3516-59. doi: 10.3390/md12063516. Mar Drugs. 2014. PMID: 24918453 Free PMC article. Review. - Linking Biosynthetic Gene Clusters to their Metabolites via Pathway- Targeted Molecular Networking.
Trautman EP, Crawford JM. Trautman EP, et al. Curr Top Med Chem. 2016;16(15):1705-16. doi: 10.2174/1568026616666151012111046. Curr Top Med Chem. 2016. PMID: 26456470 Free PMC article. Review. - Natural products: a continuing source of novel drug leads.
Cragg GM, Newman DJ. Cragg GM, et al. Biochim Biophys Acta. 2013 Jun;1830(6):3670-95. doi: 10.1016/j.bbagen.2013.02.008. Epub 2013 Feb 18. Biochim Biophys Acta. 2013. PMID: 23428572 Free PMC article. Review. - Microbial genome mining answers longstanding biosynthetic questions.
Crawford JM, Clardy J. Crawford JM, et al. Proc Natl Acad Sci U S A. 2012 May 15;109(20):7589-90. doi: 10.1073/pnas.1205361109. Epub 2012 May 1. Proc Natl Acad Sci U S A. 2012. PMID: 22550177 Free PMC article. No abstract available. - Environmental DNA-encoded antibiotics fasamycins A and B inhibit FabF in type II fatty acid biosynthesis.
Feng Z, Chakraborty D, Dewell SB, Reddy BV, Brady SF. Feng Z, et al. J Am Chem Soc. 2012 Feb 15;134(6):2981-7. doi: 10.1021/ja207662w. Epub 2012 Feb 2. J Am Chem Soc. 2012. PMID: 22224500 Free PMC article.
References
- Gans J, Wolinsky M, Dunbar J. Science. 2005;309:1387–1390. - PubMed
- Torsvik V, Ovreas L, Thingstad TF. Science. 2002;296:1064–1066. - PubMed
- Jannasch HW, Jones GE. Limnol Oceanography. 1959;4:128–139.
- Kaeberlein T, Lewis K, Epstein SS. Science. 2002;296:1127–1129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM071425/GM/NIGMS NIH HHS/United States
- R01 GM071425-01A1/GM/NIGMS NIH HHS/United States
- GM 077516/GM/NIGMS NIH HHS/United States
- GM 071425/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous