Image segmentation and dynamic lineage analysis in single-cell fluorescence microscopy - PubMed (original) (raw)
Comparative Study
Image segmentation and dynamic lineage analysis in single-cell fluorescence microscopy
Quanli Wang et al. Cytometry A. 2010 Jan.
Abstract
An increasingly common component of studies in synthetic and systems biology is analysis of dynamics of gene expression at the single-cell level, a context that is heavily dependent on the use of time-lapse movies. Extracting quantitative data on the single-cell temporal dynamics from such movies remains a major challenge. Here, we describe novel methods for automating key steps in the analysis of single-cell, fluorescent images-segmentation and lineage reconstruction-to recognize and track individual cells over time. The automated analysis iteratively combines a set of extended morphological methods for segmentation, and uses a neighborhood-based scoring method for frame-to-frame lineage linking. Our studies with bacteria, budding yeast and human cells, demonstrate the portability and usability of these methods, whether using phase, bright field or fluorescent images. These examples also demonstrate the utility of our integrated approach in facilitating analyses of engineered and natural cellular networks in diverse settings. The automated methods are implemented in freely available, open-source software.
Figures
Figure 1
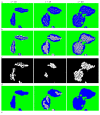
Preprocessing to convert the input grey-scale phase image into a hybrid image. (a) Sample grey-scale input images from E. coli experiment at time point t = 10,t = 25 and t = 40 respectively. (b) Resulting hybrid images after applying the modified range filter to identify initial backgrounds (colored in green). (c) Resulting hybrid images after applying the high-pass filter to (b) to further identif border regions (colored in blue). (d) the resulting hybrid images after applying a threshold filter to (c) to mark more pixels as borders.
Figure 2
Iterative segmentation. (a) Result of using a hybrid quantile filter on images in Figure 1d. (b) Hybrid images after further smoothing, thickening and re-smoothing to (a). (c) The binary masks of the blobs with lower scores in (b). (d) The blobs with higher scores in (b). (e) The final segmentation result with (arbitrary) cell coloring for visual clarity.
Figure 2
Iterative segmentation. (a) Result of using a hybrid quantile filter on images in Figure 1d. (b) Hybrid images after further smoothing, thickening and re-smoothing to (a). (c) The binary masks of the blobs with lower scores in (b). (d) The blobs with higher scores in (b). (e) The final segmentation result with (arbitrary) cell coloring for visual clarity.
Figure 3
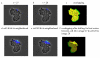
Neighborhood based cell tracking. (a,b) Two images at consecutive times with the same cell labeled in blue. (c) Result of overlaying the segmented images from a and (b). (d,e) Neighborhood of the cell identified in (a) and (b) respectively, labeled in blue and cyan. (f) The overlapping position where the best scores are obtained for the labeled cells in and (b).
Figure 4
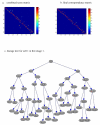
Cell tracking and lineage tree reconstruction. (a) The combined score matrix displayed as a heat map, with warm color indicating good matching. (b) Heat map for the final correspondence matrix. (c) Complete lineage tree for cell number 1 tracked in images 1-25.
Figure 5
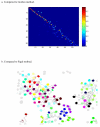
Comparison with other leading algorithms. (a) Heat map of score matrix analogous to Figure 4(a) using the method of (20). (b) Tracking result for human cells compared to that of (26).
Similar articles
- DeLTA: Automated cell segmentation, tracking, and lineage reconstruction using deep learning.
Lugagne JB, Lin H, Dunlop MJ. Lugagne JB, et al. PLoS Comput Biol. 2020 Apr 13;16(4):e1007673. doi: 10.1371/journal.pcbi.1007673. eCollection 2020 Apr. PLoS Comput Biol. 2020. PMID: 32282792 Free PMC article. - SuperSegger: robust image segmentation, analysis and lineage tracking of bacterial cells.
Stylianidou S, Brennan C, Nissen SB, Kuwada NJ, Wiggins PA. Stylianidou S, et al. Mol Microbiol. 2016 Nov;102(4):690-700. doi: 10.1111/mmi.13486. Epub 2016 Sep 23. Mol Microbiol. 2016. PMID: 27569113 - ESC-Track: A computer workflow for 4-D segmentation, tracking, lineage tracing and dynamic context analysis of ESCs.
Fernández-de-Manúel L, Díaz-Díaz C, Jiménez-Carretero D, Torres M, Montoya MC. Fernández-de-Manúel L, et al. Biotechniques. 2017 May 1;62(5):215-222. doi: 10.2144/000114545. Biotechniques. 2017. PMID: 28528574 - An integrated image analysis platform to quantify signal transduction in single cells.
Pelet S, Dechant R, Lee SS, van Drogen F, Peter M. Pelet S, et al. Integr Biol (Camb). 2012 Oct;4(10):1274-82. doi: 10.1039/c2ib20139a. Integr Biol (Camb). 2012. PMID: 22976484 - Using movies to analyse gene circuit dynamics in single cells.
Locke JC, Elowitz MB. Locke JC, et al. Nat Rev Microbiol. 2009 May;7(5):383-92. doi: 10.1038/nrmicro2056. Nat Rev Microbiol. 2009. PMID: 19369953 Free PMC article. Review.
Cited by
- Identification of individual cells from z-stacks of bright-field microscopy images.
Lugagne JB, Jain S, Ivanovitch P, Ben Meriem Z, Vulin C, Fracassi C, Batt G, Hersen P. Lugagne JB, et al. Sci Rep. 2018 Jul 30;8(1):11455. doi: 10.1038/s41598-018-29647-5. Sci Rep. 2018. PMID: 30061662 Free PMC article. - Long-term tracking of budding yeast cells in brightfield microscopy: CellStar and the Evaluation Platform.
Versari C, Stoma S, Batmanov K, Llamosi A, Mroz F, Kaczmarek A, Deyell M, Lhoussaine C, Hersen P, Batt G. Versari C, et al. J R Soc Interface. 2017 Feb;14(127):20160705. doi: 10.1098/rsif.2016.0705. J R Soc Interface. 2017. PMID: 28179544 Free PMC article. - Robust cell tracking in epithelial tissues through identification of maximum common subgraphs.
Kursawe J, Bardenet R, Zartman JJ, Baker RE, Fletcher AG. Kursawe J, et al. J R Soc Interface. 2016 Nov;13(124):20160725. doi: 10.1098/rsif.2016.0725. J R Soc Interface. 2016. PMID: 28334699 Free PMC article. - GoIFISH: a system for the quantification of single cell heterogeneity from IFISH images.
Trinh A, Rye IH, Almendro V, Helland A, Russnes HG, Markowetz F. Trinh A, et al. Genome Biol. 2014 Aug 26;15(8):442. doi: 10.1186/s13059-014-0442-y. Genome Biol. 2014. PMID: 25168174 Free PMC article. - A convolutional neural network for segmentation of yeast cells without manual training annotations.
Kruitbosch HT, Mzayek Y, Omlor S, Guerra P, Milias-Argeitis A. Kruitbosch HT, et al. Bioinformatics. 2022 Feb 7;38(5):1427-1433. doi: 10.1093/bioinformatics/btab835. Bioinformatics. 2022. PMID: 34893817 Free PMC article.
References
- Megason SG, Fraser SE. Imaging in Systems Biology. Cell. 2007;130:784–795. - PubMed
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression on a single cell. Science. 2002;297:1183–1186. - PubMed
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. - PubMed
Publication types
MeSH terms
Grants and funding
- P50 GM081883/GM/NIGMS NIH HHS/United States
- U54 CA112952/CA/NCI NIH HHS/United States
- P50-GM-081883/GM/NIGMS NIH HHS/United States
- U54-CA-112952/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases